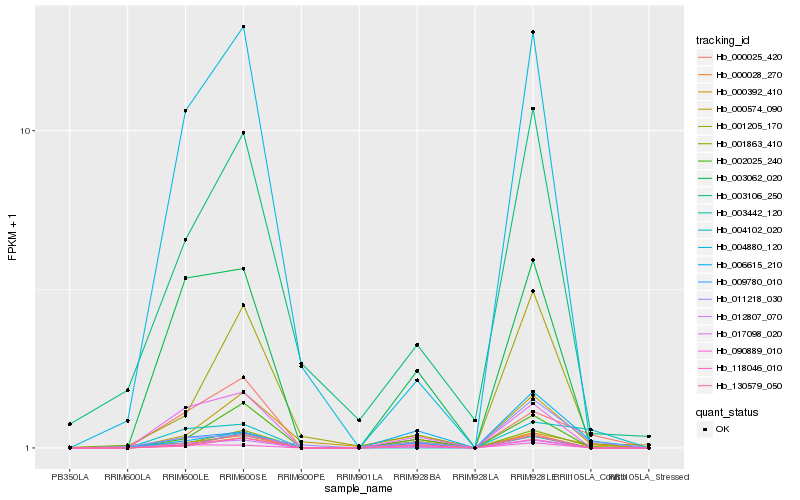
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_410 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104882159 [Vitis vinifera] |
| 2 |
Hb_000025_420 |
0.1922890516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012807_070 |
0.2240286964 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 4 |
Hb_130579_050 |
0.2302254608 |
- |
- |
hypothetical protein JCGZ_25577 [Jatropha curcas] |
| 5 |
Hb_000574_090 |
0.2352124226 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 6 |
Hb_001863_410 |
0.2383756346 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 7 |
Hb_001205_170 |
0.2433815121 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002025_240 |
0.2455543394 |
- |
- |
- |
| 9 |
Hb_004102_020 |
0.2457759763 |
- |
- |
- |
| 10 |
Hb_017098_020 |
0.253567257 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 11 |
Hb_090889_010 |
0.253700951 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 12 |
Hb_003062_020 |
0.2552696194 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 13 |
Hb_004880_120 |
0.2588773361 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 14 |
Hb_011218_030 |
0.2603514411 |
- |
- |
hypothetical protein VITISV_002891 [Vitis vinifera] |
| 15 |
Hb_118046_010 |
0.2604478746 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 16 |
Hb_000028_270 |
0.2612316376 |
- |
- |
- |
| 17 |
Hb_009780_010 |
0.2623059512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003442_120 |
0.2643633221 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 19 |
Hb_006615_210 |
0.2649900753 |
- |
- |
PREDICTED: uncharacterized protein LOC105640677 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003106_250 |
0.2651891608 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |