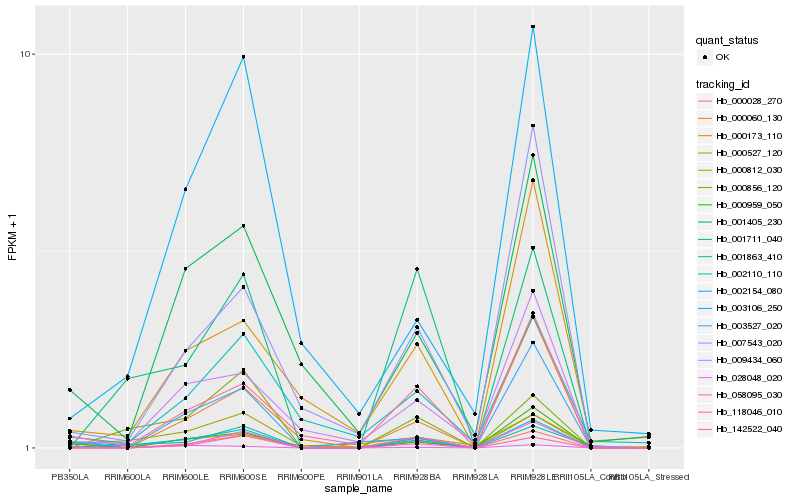
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002154_080 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a018017mg, partial [Erythranthe guttata] |
| 2 |
Hb_000959_050 |
0.1922101523 |
- |
- |
PREDICTED: uncharacterized protein LOC102627366 [Citrus sinensis] |
| 3 |
Hb_001863_410 |
0.1995537285 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 4 |
Hb_142522_040 |
0.212876015 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_000812_030 |
0.2396681817 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 6 |
Hb_000028_270 |
0.2425626852 |
- |
- |
- |
| 7 |
Hb_000527_120 |
0.2462200882 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 8 |
Hb_001405_230 |
0.2477179301 |
- |
- |
STELAR K+ outward rectifier isoform 2 [Theobroma cacao] |
| 9 |
Hb_028048_020 |
0.2477830854 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 10 |
Hb_001711_040 |
0.2489517665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_009434_060 |
0.2490726111 |
- |
- |
T4.15 [Malus x robusta] |
| 12 |
Hb_000060_130 |
0.2513263084 |
- |
- |
- |
| 13 |
Hb_003527_020 |
0.2522633439 |
- |
- |
BnaC01g27150D [Brassica napus] |
| 14 |
Hb_000856_120 |
0.2585996503 |
transcription factor |
TF Family: LOB |
PREDICTED: myb-like protein M [Pyrus x bretschneideri] |
| 15 |
Hb_058095_030 |
0.2628324559 |
- |
- |
- |
| 16 |
Hb_002110_110 |
0.2688008167 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 17 |
Hb_118046_010 |
0.2693446421 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 18 |
Hb_007543_020 |
0.2696182835 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 19 |
Hb_003106_250 |
0.2698676038 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 20 |
Hb_000173_110 |
0.2699763362 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |