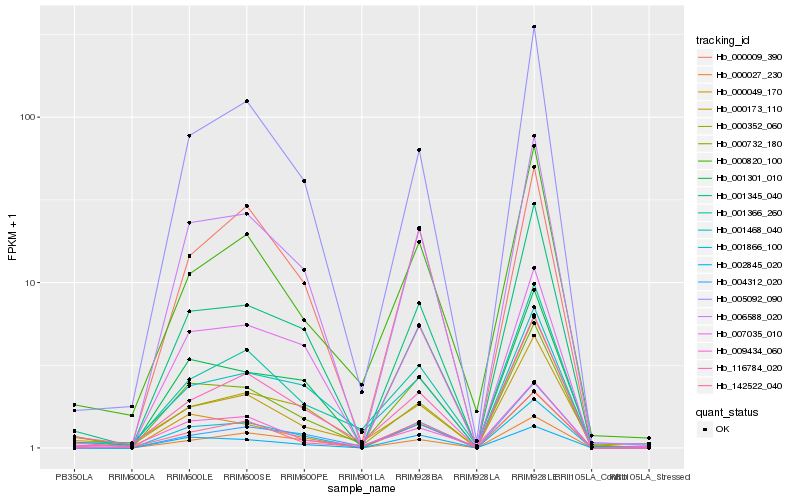
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000732_180 |
0.0 |
- |
- |
Uncharacterized protein TCM_030007 [Theobroma cacao] |
| 2 |
Hb_142522_040 |
0.1086855922 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_001345_040 |
0.1254880651 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 4 |
Hb_116784_020 |
0.1258680904 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_006588_020 |
0.1269270849 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_001366_260 |
0.1327201772 |
- |
- |
T4.15 [Malus x robusta] |
| 7 |
Hb_002845_020 |
0.1338523227 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 8 |
Hb_001866_100 |
0.1339315294 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 9 |
Hb_000049_170 |
0.1360716621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_009434_060 |
0.1371976447 |
- |
- |
T4.15 [Malus x robusta] |
| 11 |
Hb_000820_100 |
0.1395709517 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 12 |
Hb_001468_040 |
0.1424920833 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 13 |
Hb_000173_110 |
0.1457586236 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 14 |
Hb_005092_090 |
0.1501739137 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 15 |
Hb_001301_010 |
0.1512059457 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000027_230 |
0.1553775858 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 17 |
Hb_004312_020 |
0.1564697507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007035_010 |
0.1568092471 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000352_060 |
0.1589073731 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 20 |
Hb_000009_390 |
0.1592848648 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |