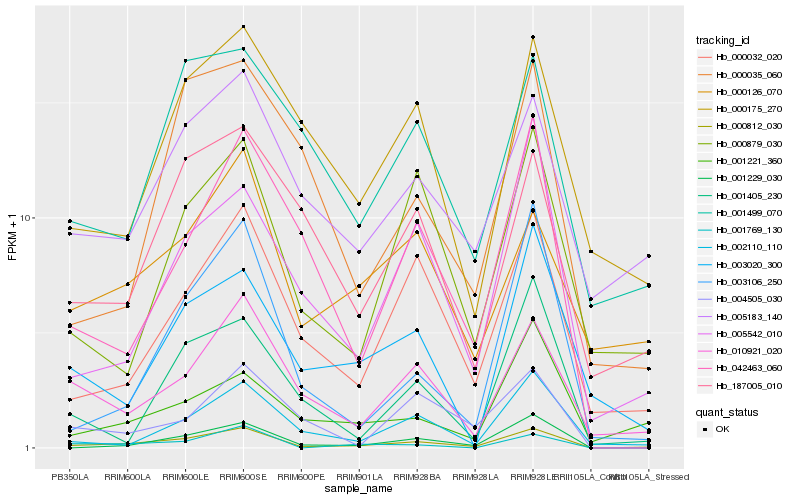
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000812_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 2 |
Hb_000032_020 |
0.1792055749 |
- |
- |
Centromeric protein E, putative [Ricinus communis] |
| 3 |
Hb_010921_020 |
0.1871099892 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 4 |
Hb_001229_030 |
0.1877458626 |
- |
- |
PREDICTED: uncharacterized protein LOC104783713 [Camelina sativa] |
| 5 |
Hb_002110_110 |
0.1970060245 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 6 |
Hb_003106_250 |
0.1990623926 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 7 |
Hb_005183_140 |
0.2010549876 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 8 |
Hb_000879_030 |
0.2019182327 |
- |
- |
PREDICTED: uncharacterized protein LOC103707552 [Phoenix dactylifera] |
| 9 |
Hb_005542_010 |
0.2033148693 |
- |
- |
Uncharacterized protein TCM_021447 [Theobroma cacao] |
| 10 |
Hb_001769_130 |
0.205522895 |
- |
- |
- |
| 11 |
Hb_042463_060 |
0.2081844959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000126_070 |
0.2087875598 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Gossypium arboreum] |
| 13 |
Hb_000175_270 |
0.2163281395 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 14 |
Hb_187005_010 |
0.2175304202 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 15 |
Hb_001405_230 |
0.220875087 |
- |
- |
STELAR K+ outward rectifier isoform 2 [Theobroma cacao] |
| 16 |
Hb_001499_070 |
0.2212505369 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 17 |
Hb_001221_360 |
0.2217504059 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 18 |
Hb_003020_300 |
0.2238183209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004505_030 |
0.2305227598 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_000035_060 |
0.2323714103 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |