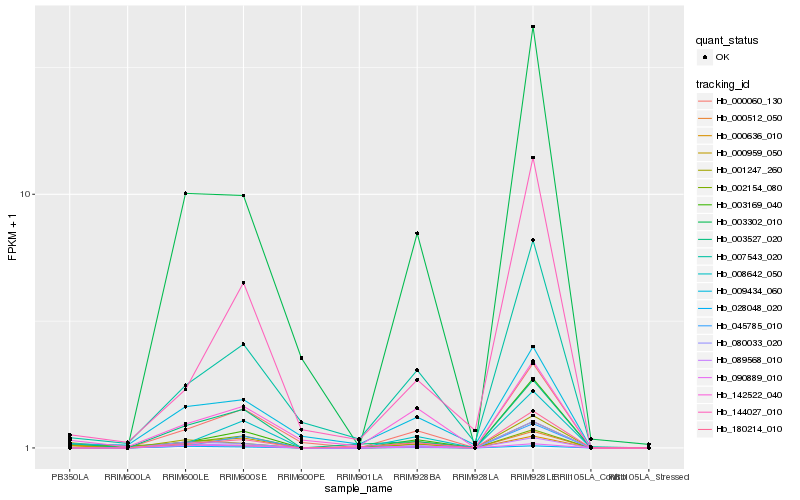
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000959_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102627366 [Citrus sinensis] |
| 2 |
Hb_000060_130 |
0.1506768628 |
- |
- |
- |
| 3 |
Hb_003527_020 |
0.1525733698 |
- |
- |
BnaC01g27150D [Brassica napus] |
| 4 |
Hb_001247_260 |
0.1615108088 |
- |
- |
- |
| 5 |
Hb_000636_010 |
0.1621603266 |
- |
- |
PREDICTED: uncharacterized protein LOC105767086 [Gossypium raimondii] |
| 6 |
Hb_002154_080 |
0.1922101523 |
- |
- |
hypothetical protein MIMGU_mgv1a018017mg, partial [Erythranthe guttata] |
| 7 |
Hb_007543_020 |
0.2003442694 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 8 |
Hb_142522_040 |
0.2019099725 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_003302_010 |
0.2083946824 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 10 |
Hb_000512_050 |
0.2095570479 |
- |
- |
hypothetical protein VITISV_005640 [Vitis vinifera] |
| 11 |
Hb_080033_020 |
0.2111749527 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 12 |
Hb_008642_050 |
0.2149508349 |
- |
- |
- |
| 13 |
Hb_089568_010 |
0.2163801159 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 14 |
Hb_009434_060 |
0.2173161223 |
- |
- |
T4.15 [Malus x robusta] |
| 15 |
Hb_028048_020 |
0.2195880717 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 16 |
Hb_180214_010 |
0.2200669814 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 17 |
Hb_045785_010 |
0.2211038901 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 18 |
Hb_144027_010 |
0.2220551029 |
- |
- |
- |
| 19 |
Hb_003169_040 |
0.2240608674 |
- |
- |
- |
| 20 |
Hb_090889_010 |
0.2288731618 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |