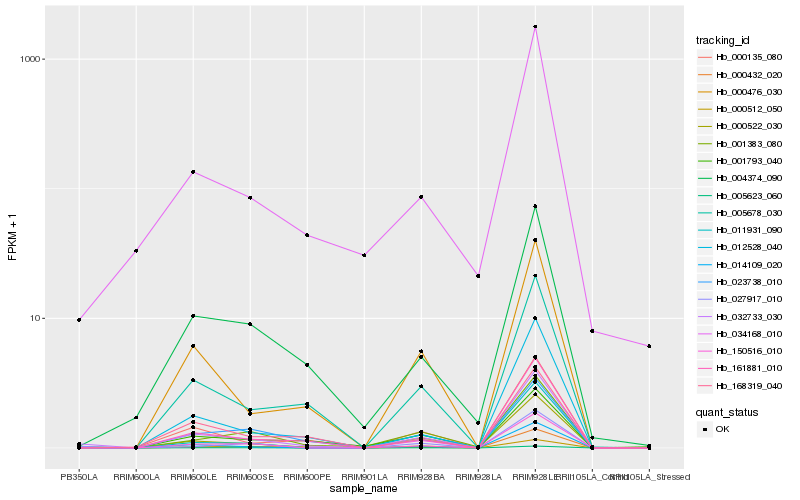
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032733_030 |
0.0 |
- |
- |
hypothetical protein CISIN_1g048657mg, partial [Citrus sinensis] |
| 2 |
Hb_000522_030 |
0.1224792403 |
- |
- |
hypothetical protein VITISV_006017 [Vitis vinifera] |
| 3 |
Hb_012528_040 |
0.1314070633 |
- |
- |
- |
| 4 |
Hb_014109_020 |
0.1338902001 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 5 |
Hb_000135_080 |
0.1410726944 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 6 |
Hb_005623_060 |
0.1411510416 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_005678_030 |
0.1437272116 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 8 |
Hb_000476_030 |
0.144804235 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 9 |
Hb_027917_010 |
0.1461474533 |
- |
- |
PREDICTED: uncharacterized protein LOC103699089, partial [Phoenix dactylifera] |
| 10 |
Hb_000432_020 |
0.1528408966 |
desease resistance |
Gene Name: Tropomyosin_1 |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_011931_090 |
0.1577733844 |
- |
- |
- |
| 12 |
Hb_150516_010 |
0.1651731163 |
- |
- |
PREDICTED: uncharacterized protein LOC104886268, partial [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_001383_080 |
0.165247886 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |
| 14 |
Hb_000512_050 |
0.1670703165 |
- |
- |
hypothetical protein VITISV_005640 [Vitis vinifera] |
| 15 |
Hb_034168_010 |
0.168548009 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 16 |
Hb_001793_040 |
0.1693606826 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 17 |
Hb_161881_010 |
0.1711331569 |
- |
- |
laccase, putative [Ricinus communis] |
| 18 |
Hb_168319_040 |
0.1738350121 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 19 |
Hb_004374_090 |
0.1740164349 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 20 |
Hb_023738_010 |
0.1759219942 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |