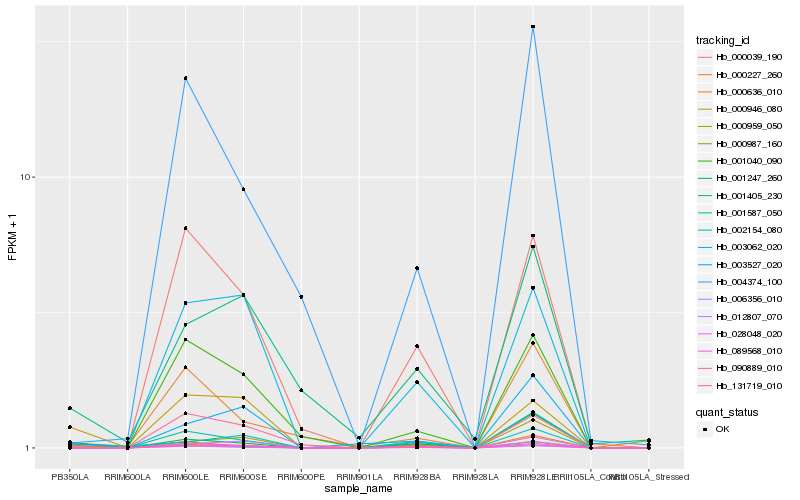
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028048_020 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 2 |
Hb_000636_010 |
0.2173971237 |
- |
- |
PREDICTED: uncharacterized protein LOC105767086 [Gossypium raimondii] |
| 3 |
Hb_000946_080 |
0.2175899064 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Prunus mume] |
| 4 |
Hb_000959_050 |
0.2195880717 |
- |
- |
PREDICTED: uncharacterized protein LOC102627366 [Citrus sinensis] |
| 5 |
Hb_090889_010 |
0.2260162254 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 6 |
Hb_000039_190 |
0.2394650949 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 7 |
Hb_001247_260 |
0.2412078851 |
- |
- |
- |
| 8 |
Hb_006356_010 |
0.2441107849 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 9 |
Hb_003062_020 |
0.2445764654 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 10 |
Hb_002154_080 |
0.2477830854 |
- |
- |
hypothetical protein MIMGU_mgv1a018017mg, partial [Erythranthe guttata] |
| 11 |
Hb_131719_010 |
0.250025615 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 12 |
Hb_001040_090 |
0.2530117836 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |
| 13 |
Hb_003527_020 |
0.2553965577 |
- |
- |
BnaC01g27150D [Brassica napus] |
| 14 |
Hb_089568_010 |
0.2592084387 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 15 |
Hb_000227_260 |
0.2613289461 |
- |
- |
PREDICTED: RAN GTPase-activating protein 2 [Jatropha curcas] |
| 16 |
Hb_001587_050 |
0.2615319679 |
- |
- |
- |
| 17 |
Hb_001405_230 |
0.2624577845 |
- |
- |
STELAR K+ outward rectifier isoform 2 [Theobroma cacao] |
| 18 |
Hb_000987_160 |
0.2683212411 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 19 |
Hb_012807_070 |
0.2715743212 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 20 |
Hb_004374_100 |
0.2734476761 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |