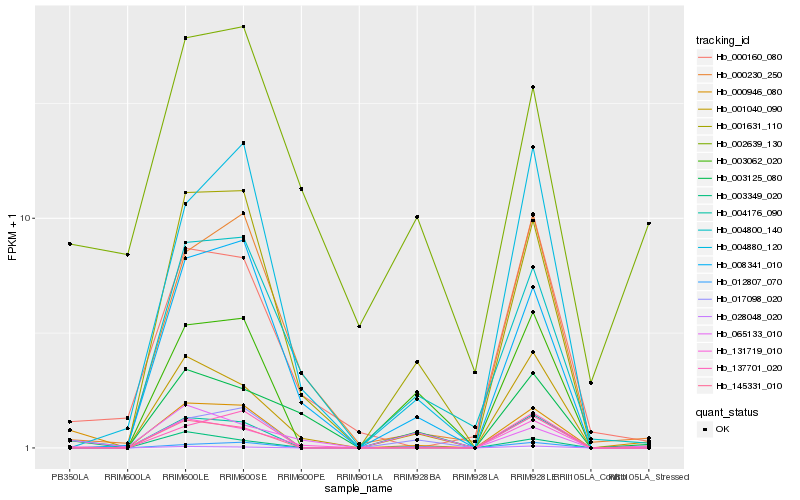
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000946_080 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Prunus mume] |
| 2 |
Hb_028048_020 |
0.2175899064 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 3 |
Hb_131719_010 |
0.2625508797 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 4 |
Hb_002639_130 |
0.2872043883 |
- |
- |
hypothetical protein JCGZ_08129 [Jatropha curcas] |
| 5 |
Hb_004176_090 |
0.2885799072 |
- |
- |
Nuc-1 negative regulatory protein preg, putative [Ricinus communis] |
| 6 |
Hb_001631_110 |
0.2909444745 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: gamma-glutamyltranspeptidase 1-like [Populus euphratica] |
| 7 |
Hb_017098_020 |
0.2968813837 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 8 |
Hb_145331_010 |
0.2977048138 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0016s07690g [Populus trichocarpa] |
| 9 |
Hb_000230_250 |
0.2990724302 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000160_080 |
0.3014684706 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM2b-like [Jatropha curcas] |
| 11 |
Hb_003125_080 |
0.304309455 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 12 |
Hb_004800_140 |
0.3045809999 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003062_020 |
0.3046525026 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 14 |
Hb_008341_010 |
0.3060959604 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_137701_020 |
0.3074925534 |
- |
- |
putative protein kinase [Oryza sativa Japonica Group] |
| 16 |
Hb_003349_020 |
0.3080081808 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_065133_010 |
0.3098971533 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 18 |
Hb_012807_070 |
0.3099261688 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 19 |
Hb_001040_090 |
0.31286174 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |
| 20 |
Hb_004880_120 |
0.3135469421 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |