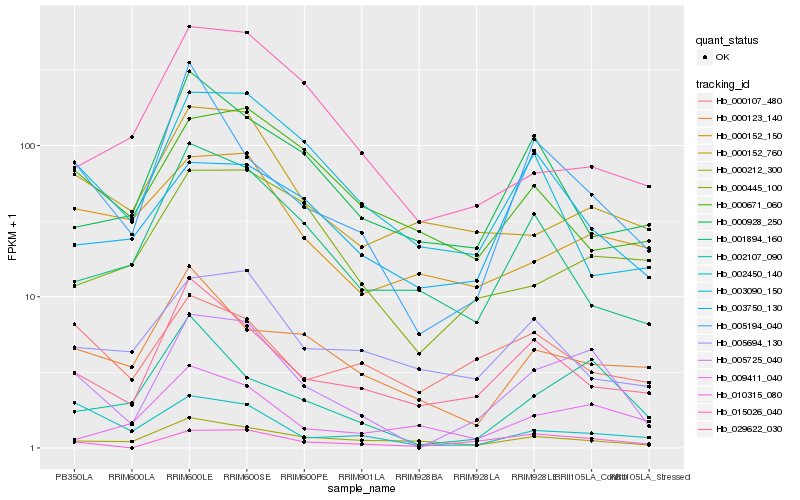
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005725_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_002107_090 |
0.2154063558 |
transcription factor |
TF Family: bZIP |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 3 |
Hb_010315_080 |
0.2169784181 |
- |
- |
- |
| 4 |
Hb_029622_030 |
0.2323965788 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_005194_040 |
0.2430592565 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000152_760 |
0.2440190384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000212_300 |
0.2459267791 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 8 |
Hb_002450_140 |
0.2489682767 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 9 |
Hb_000152_150 |
0.2515168941 |
- |
- |
PREDICTED: uncharacterized protein LOC105648558 [Jatropha curcas] |
| 10 |
Hb_000445_100 |
0.2540275474 |
- |
- |
PREDICTED: glutathione S-transferase TCHQD [Jatropha curcas] |
| 11 |
Hb_003090_150 |
0.2600513987 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 12 |
Hb_005694_130 |
0.2617911451 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 13 |
Hb_001894_160 |
0.2623967046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009411_040 |
0.2630725498 |
- |
- |
- |
| 15 |
Hb_000123_140 |
0.2638056941 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 16 |
Hb_003750_130 |
0.2675318398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000107_480 |
0.2705363469 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_015026_040 |
0.2706317642 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 19 |
Hb_000928_250 |
0.2707591211 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_000671_060 |
0.2708681858 |
- |
- |
protein binding protein, putative [Ricinus communis] |