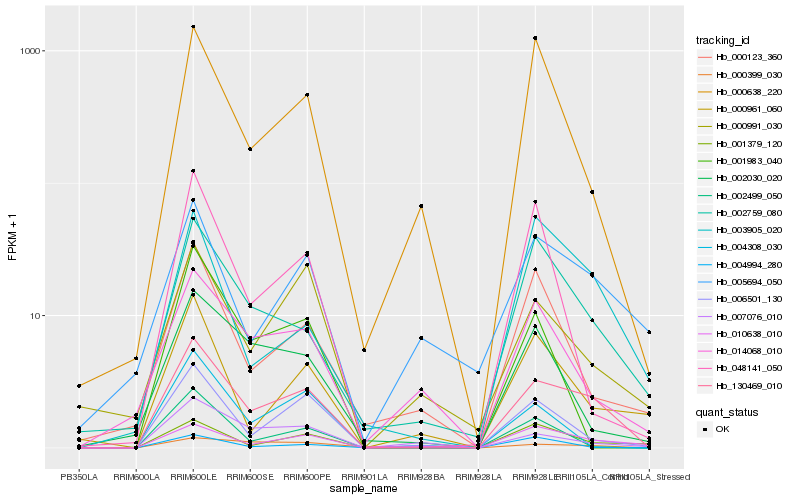
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_130469_010 |
0.0 |
- |
- |
pleiotropic drug resistance ABC transporter family protein, partial [Eleutherococcus senticosus] |
| 2 |
Hb_006501_130 |
0.2101881961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002759_080 |
0.2212416668 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 4 |
Hb_010638_010 |
0.2247835007 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 5 |
Hb_002499_050 |
0.2372075094 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_001379_120 |
0.2374205566 |
- |
- |
hypothetical protein POPTR_0001s29730g [Populus trichocarpa] |
| 7 |
Hb_000638_220 |
0.2391208962 |
- |
- |
PREDICTED: photosystem I reaction center subunit N, chloroplastic [Jatropha curcas] |
| 8 |
Hb_014068_010 |
0.2405993027 |
- |
- |
PREDICTED: phototropin-1 [Jatropha curcas] |
| 9 |
Hb_004994_280 |
0.2447111182 |
- |
- |
hypothetical protein JCGZ_14421 [Jatropha curcas] |
| 10 |
Hb_007076_010 |
0.244750083 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_005694_050 |
0.2457337856 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 12 |
Hb_000961_060 |
0.2467603201 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 13 |
Hb_002030_020 |
0.2472333555 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 14 |
Hb_000991_030 |
0.2489169339 |
- |
- |
PREDICTED: uncharacterized protein LOC105122436 [Populus euphratica] |
| 15 |
Hb_000123_360 |
0.2523692052 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_001983_040 |
0.2525870956 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 17 |
Hb_048141_050 |
0.253281118 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 18 |
Hb_000399_030 |
0.2537109102 |
- |
- |
PREDICTED: uncharacterized protein LOC105634579 [Jatropha curcas] |
| 19 |
Hb_004308_030 |
0.2544692412 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 20 |
Hb_003905_020 |
0.2545618658 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |