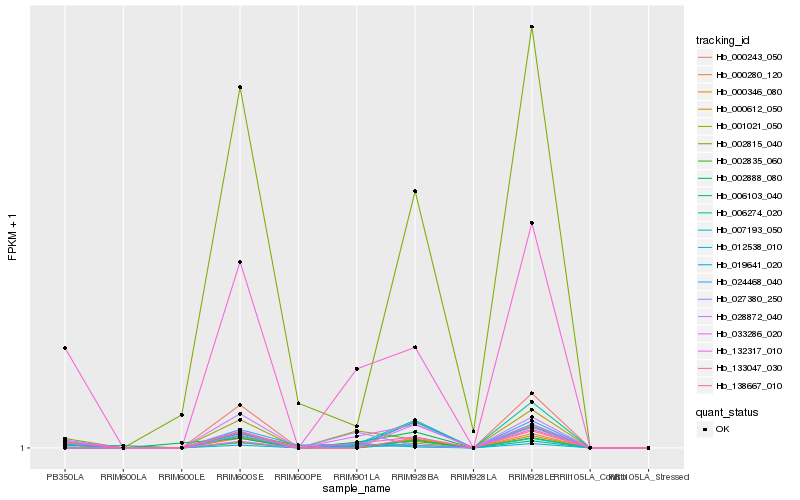
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_133047_030 |
0.0 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_132317_010 |
0.0938731888 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_000346_080 |
0.2119631242 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 4 |
Hb_012538_010 |
0.2245800115 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_006103_040 |
0.2290694457 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_033286_020 |
0.2546864316 |
- |
- |
- |
| 7 |
Hb_002888_080 |
0.2564710613 |
- |
- |
PREDICTED: uncharacterized protein LOC104120414 [Nicotiana tomentosiformis] |
| 8 |
Hb_007193_050 |
0.2611262349 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 9 |
Hb_024468_040 |
0.2673961191 |
- |
- |
- |
| 10 |
Hb_002835_060 |
0.2730514336 |
- |
- |
PREDICTED: uncharacterized protein LOC105640295 [Jatropha curcas] |
| 11 |
Hb_001021_050 |
0.2796426905 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 12 |
Hb_019641_020 |
0.2879433436 |
- |
- |
hypothetical protein AALP_AAs49670U000100, partial [Arabis alpina] |
| 13 |
Hb_006274_020 |
0.2897481214 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 14 |
Hb_002815_040 |
0.2909698097 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 15 |
Hb_000612_050 |
0.2983685937 |
- |
- |
hypothetical protein MIMGU_mgv1a020753mg, partial [Erythranthe guttata] |
| 16 |
Hb_138667_010 |
0.3011140274 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 17 |
Hb_000280_120 |
0.3028612951 |
- |
- |
PREDICTED: uncharacterized protein LOC105643934 [Jatropha curcas] |
| 18 |
Hb_000243_050 |
0.3036212683 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 19 |
Hb_027380_250 |
0.3043805549 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 20 |
Hb_028872_040 |
0.3045187438 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |