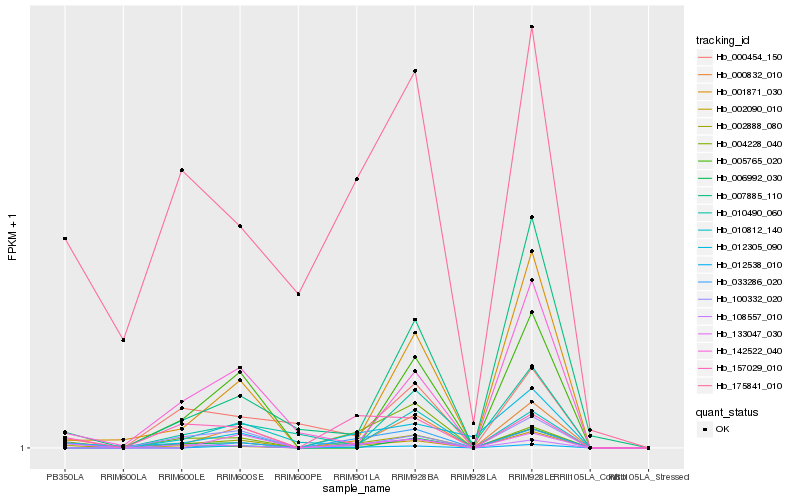
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002888_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104120414 [Nicotiana tomentosiformis] |
| 2 |
Hb_157029_010 |
0.2176440066 |
- |
- |
PREDICTED: uncharacterized protein LOC101497268 [Cicer arietinum] |
| 3 |
Hb_108557_010 |
0.2203685846 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 4 |
Hb_004228_040 |
0.2226696923 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 5 |
Hb_100332_020 |
0.2324989616 |
- |
- |
PREDICTED: uncharacterized protein LOC105168695 [Sesamum indicum] |
| 6 |
Hb_000832_010 |
0.2337440714 |
- |
- |
hypothetical protein VITISV_005044 [Vitis vinifera] |
| 7 |
Hb_001871_030 |
0.2344495485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002090_010 |
0.2362363119 |
- |
- |
gag-pol fusion protein [Rhizophagus irregularis DAOM 197198w] |
| 9 |
Hb_142522_040 |
0.2431194671 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_033286_020 |
0.2476086692 |
- |
- |
- |
| 11 |
Hb_000454_150 |
0.2495965156 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 12 |
Hb_012305_090 |
0.2512809862 |
- |
- |
hypothetical protein VITISV_018166 [Vitis vinifera] |
| 13 |
Hb_175841_010 |
0.2540171601 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 14 |
Hb_133047_030 |
0.2564710613 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_007885_110 |
0.2613565531 |
- |
- |
PREDICTED: uncharacterized protein LOC104109376 [Nicotiana tomentosiformis] |
| 16 |
Hb_010812_140 |
0.2638752581 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_005765_020 |
0.2727191504 |
- |
- |
- |
| 18 |
Hb_012538_010 |
0.2735539069 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_010490_060 |
0.2744676547 |
- |
- |
PREDICTED: uncharacterized protein LOC105767021 [Gossypium raimondii] |
| 20 |
Hb_006992_030 |
0.276538425 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |