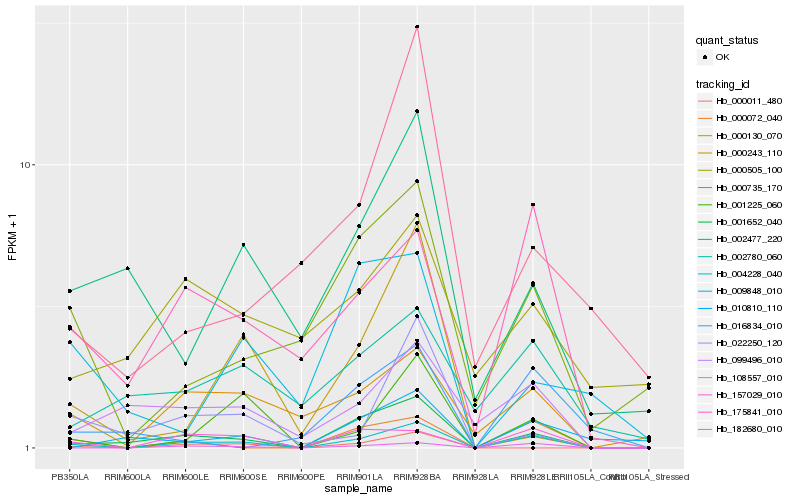
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001652_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103341683 [Prunus mume] |
| 2 |
Hb_004228_040 |
0.2047605135 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 3 |
Hb_099496_010 |
0.2794486701 |
- |
- |
PREDICTED: uncharacterized protein LOC105138000 [Populus euphratica] |
| 4 |
Hb_010810_110 |
0.280554215 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 5 |
Hb_002477_220 |
0.2873611126 |
- |
- |
hypothetical protein POPTR_0017s06650g [Populus trichocarpa] |
| 6 |
Hb_108557_010 |
0.2908364922 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 7 |
Hb_157029_010 |
0.3030826271 |
- |
- |
PREDICTED: uncharacterized protein LOC101497268 [Cicer arietinum] |
| 8 |
Hb_000072_040 |
0.3137920912 |
- |
- |
- |
| 9 |
Hb_000011_480 |
0.3189418574 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000735_170 |
0.3225324241 |
- |
- |
hypothetical protein JCGZ_15146 [Jatropha curcas] |
| 11 |
Hb_000243_110 |
0.3225921593 |
- |
- |
PREDICTED: WAT1-related protein At3g53210-like [Jatropha curcas] |
| 12 |
Hb_022250_120 |
0.3228610748 |
- |
- |
PREDICTED: uncharacterized protein LOC105648689 [Jatropha curcas] |
| 13 |
Hb_002780_060 |
0.3254224066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_182680_010 |
0.3289680769 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 15 |
Hb_009848_010 |
0.3298724018 |
- |
- |
putative acyl-activating enzyme 16, chloroplastic -like protein [Gossypium arboreum] |
| 16 |
Hb_000130_070 |
0.33211767 |
- |
- |
hypothetical protein JCGZ_14738 [Jatropha curcas] |
| 17 |
Hb_175841_010 |
0.3355796871 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 18 |
Hb_016834_010 |
0.3394338825 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 19 |
Hb_001225_060 |
0.3400643045 |
- |
- |
BnaC03g58850D [Brassica napus] |
| 20 |
Hb_000505_100 |
0.3466651671 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC100250663 [Vitis vinifera] |