| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
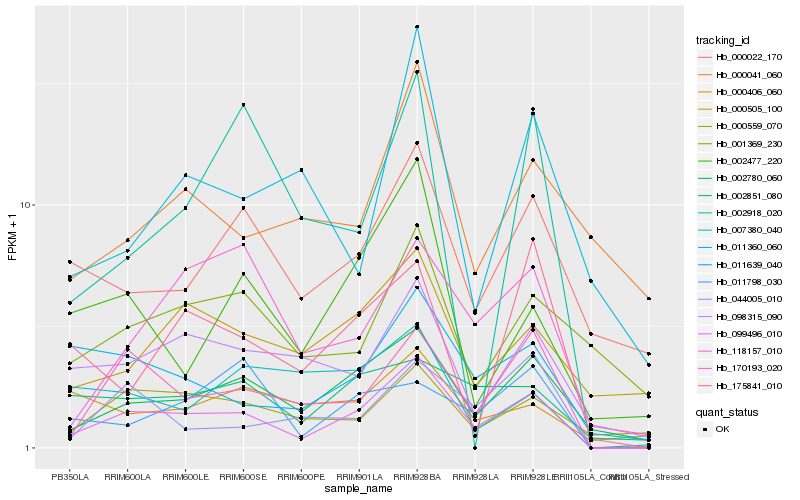
Hb_099496_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105138000 [Populus euphratica] |
| 2 |
Hb_002780_060 |
0.1765761895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_170193_020 |
0.1799705588 |
- |
- |
- |
| 4 |
Hb_098315_090 |
0.2201725153 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 5 |
Hb_011639_040 |
0.2210710682 |
- |
- |
hypothetical protein M569_08265, partial [Genlisea aurea] |
| 6 |
Hb_000022_170 |
0.232206136 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 7 |
Hb_044005_010 |
0.2342405647 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 8 |
Hb_118157_010 |
0.2352979796 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000559_070 |
0.2355535231 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 10 |
Hb_002477_220 |
0.2369010703 |
- |
- |
hypothetical protein POPTR_0017s06650g [Populus trichocarpa] |
| 11 |
Hb_002851_080 |
0.2377495713 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 12 |
Hb_000505_100 |
0.2430073067 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC100250663 [Vitis vinifera] |
| 13 |
Hb_011798_030 |
0.2479161573 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Vitis vinifera] |
| 14 |
Hb_175841_010 |
0.2520871149 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 15 |
Hb_001369_230 |
0.2536053249 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 16 |
Hb_000041_060 |
0.2595186768 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 17 |
Hb_002918_020 |
0.2612902039 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007380_040 |
0.2621541811 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_000406_060 |
0.2628456305 |
- |
- |
hypothetical protein CICLE_v10000115mg [Citrus clementina] |
| 20 |
Hb_011360_060 |
0.2655047717 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |