| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
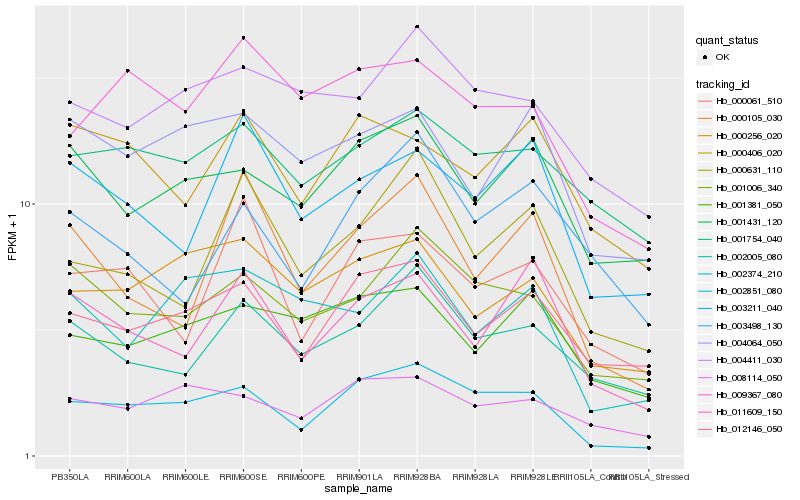
Hb_002851_080 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 2 |
Hb_001006_340 |
0.1292219554 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 3 |
Hb_008114_050 |
0.1344078504 |
- |
- |
hypothetical protein POPTR_0006s02430g [Populus trichocarpa] |
| 4 |
Hb_012146_050 |
0.1355996569 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001431_120 |
0.1483113321 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 6 |
Hb_000256_020 |
0.1498343238 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 7 |
Hb_004411_030 |
0.155101974 |
- |
- |
hypothetical protein POPTR_0017s00310g [Populus trichocarpa] |
| 8 |
Hb_009367_080 |
0.1552489723 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 42 [Jatropha curcas] |
| 9 |
Hb_000631_110 |
0.1567248978 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 10 |
Hb_002005_080 |
0.1573536507 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 11 |
Hb_011609_150 |
0.1578146316 |
- |
- |
PREDICTED: HEAT repeat-containing protein 5B [Jatropha curcas] |
| 12 |
Hb_001754_040 |
0.1604188996 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 13 |
Hb_000061_510 |
0.1606722305 |
- |
- |
hypothetical protein JCGZ_11686 [Jatropha curcas] |
| 14 |
Hb_004064_050 |
0.1640405151 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 15 |
Hb_001381_050 |
0.1650135415 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 16 |
Hb_000105_030 |
0.1660258591 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 17 |
Hb_000406_020 |
0.1664676425 |
- |
- |
- |
| 18 |
Hb_002374_210 |
0.1671018327 |
- |
- |
PREDICTED: uncharacterized protein LOC105638976 [Jatropha curcas] |
| 19 |
Hb_003211_040 |
0.1676426877 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003498_130 |
0.1683137817 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3 homolog 2-like [Citrus sinensis] |