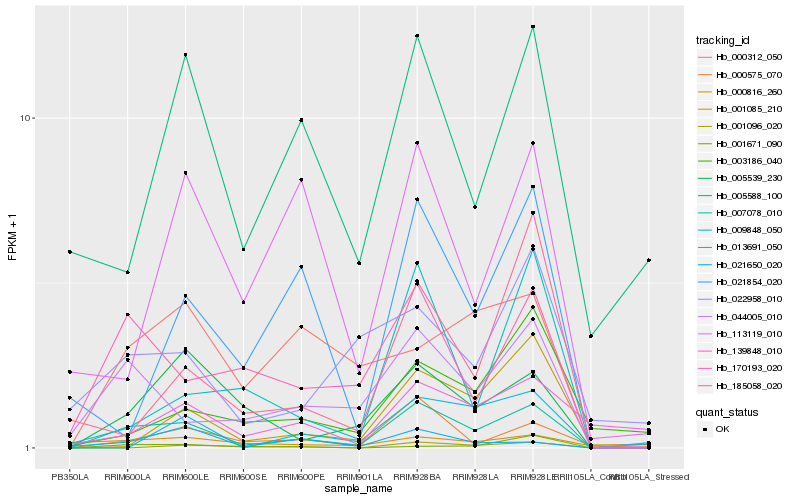
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007078_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_044005_010 |
0.2342882936 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 3 |
Hb_000575_070 |
0.2399258962 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X1 [Populus euphratica] |
| 4 |
Hb_005539_230 |
0.2518335915 |
- |
- |
hypothetical protein L484_027799 [Morus notabilis] |
| 5 |
Hb_170193_020 |
0.2535502758 |
- |
- |
- |
| 6 |
Hb_001085_210 |
0.2772083982 |
- |
- |
PREDICTED: myosin-binding protein 3 [Jatropha curcas] |
| 7 |
Hb_009848_050 |
0.2866235522 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK13 [Elaeis guineensis] |
| 8 |
Hb_185058_020 |
0.2905369038 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 9 |
Hb_001096_020 |
0.2929535849 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Populus euphratica] |
| 10 |
Hb_000312_050 |
0.2934789153 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000816_260 |
0.2943535663 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_113119_010 |
0.2975505343 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_021650_020 |
0.2997009446 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 14 |
Hb_139848_010 |
0.2998329974 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS4, chloroplastic [Populus euphratica] |
| 15 |
Hb_021854_020 |
0.3011129151 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 16 |
Hb_003186_040 |
0.3018785755 |
- |
- |
PREDICTED: protein DENND6A [Jatropha curcas] |
| 17 |
Hb_022958_010 |
0.3020290051 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 18 |
Hb_013691_050 |
0.3033759652 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 19 |
Hb_005588_100 |
0.3035386711 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 20 |
Hb_001671_090 |
0.3062942992 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |