| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
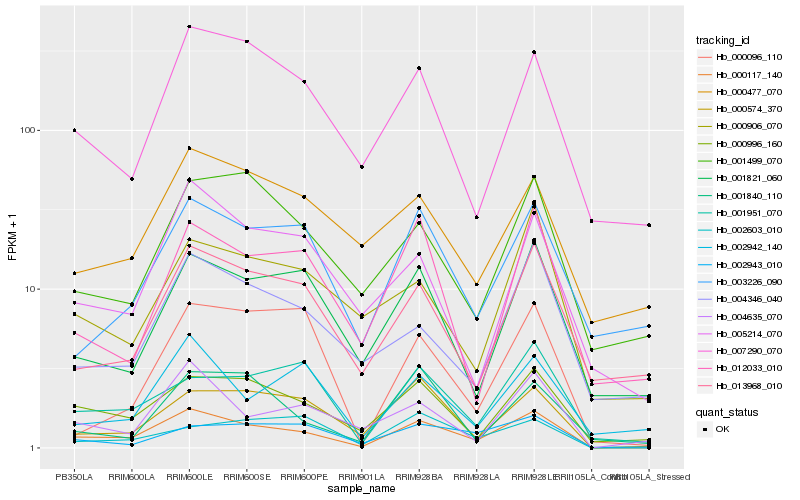
Hb_000117_140 |
0.0 |
- |
- |
hypothetical protein CISIN_1g042884mg, partial [Citrus sinensis] |
| 2 |
Hb_000996_160 |
0.1687953297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000574_370 |
0.1820265265 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 4 |
Hb_001951_070 |
0.1921868691 |
- |
- |
putative protein [Arabidopsis thaliana] |
| 5 |
Hb_004635_070 |
0.1932423013 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Solanum lycopersicum] |
| 6 |
Hb_007290_070 |
0.1940256048 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 7 |
Hb_001821_060 |
0.1980417566 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 8 |
Hb_005214_070 |
0.1991820906 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 9 |
Hb_013968_010 |
0.2000045313 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 10 |
Hb_002943_010 |
0.2062871731 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 11 |
Hb_012033_010 |
0.2068971741 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 12 |
Hb_001499_070 |
0.2095760502 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 13 |
Hb_000906_070 |
0.2095994923 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_004346_040 |
0.2100593696 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 15 |
Hb_003226_090 |
0.2116720671 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 16 |
Hb_002942_140 |
0.2124722948 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 17 |
Hb_002603_010 |
0.2165161081 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000477_070 |
0.219014558 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 19 |
Hb_001840_110 |
0.2190618612 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 20 |
Hb_000096_110 |
0.2200835023 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |