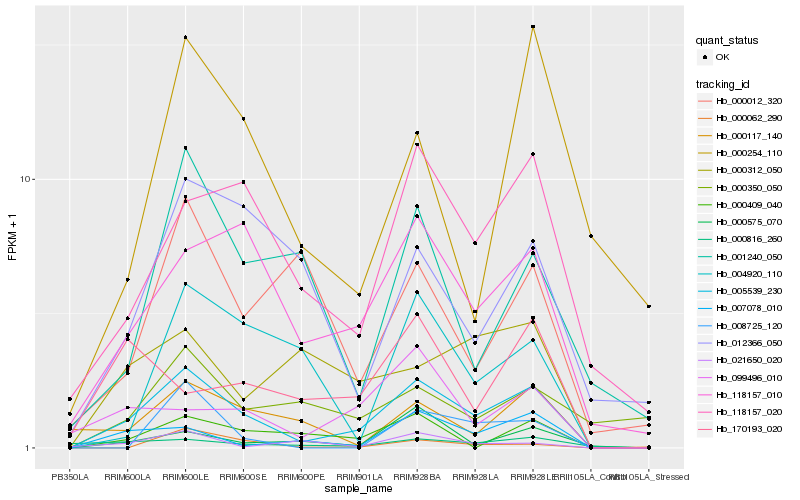
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_230 |
0.0 |
- |
- |
hypothetical protein L484_027799 [Morus notabilis] |
| 2 |
Hb_118157_010 |
0.2203416489 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 3 |
Hb_118157_020 |
0.2383698701 |
- |
- |
hypothetical protein MTR_3g456110 [Medicago truncatula] |
| 4 |
Hb_007078_010 |
0.2518335915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000409_040 |
0.2693511226 |
transcription factor |
TF Family: AP2 |
hypothetical protein POPTR_0017s11470g [Populus trichocarpa] |
| 6 |
Hb_099496_010 |
0.2696007046 |
- |
- |
PREDICTED: uncharacterized protein LOC105138000 [Populus euphratica] |
| 7 |
Hb_000117_140 |
0.2757631324 |
- |
- |
hypothetical protein CISIN_1g042884mg, partial [Citrus sinensis] |
| 8 |
Hb_000816_260 |
0.2805250731 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 9 |
Hb_000312_050 |
0.2842707899 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000575_070 |
0.2851369608 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X1 [Populus euphratica] |
| 11 |
Hb_004920_110 |
0.2864175808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_012366_050 |
0.2968161198 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 13 |
Hb_001240_050 |
0.2974057256 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Jatropha curcas] |
| 14 |
Hb_170193_020 |
0.298769996 |
- |
- |
- |
| 15 |
Hb_000012_320 |
0.2991046878 |
- |
- |
PREDICTED: uncharacterized protein LOC105638164 [Jatropha curcas] |
| 16 |
Hb_000254_110 |
0.3002753529 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 17 |
Hb_021650_020 |
0.3021631735 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 18 |
Hb_008725_120 |
0.3039349403 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Populus euphratica] |
| 19 |
Hb_000350_050 |
0.3045980906 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 20 |
Hb_000062_290 |
0.3051706022 |
desease resistance |
Gene Name: ABC_membrane |
hypothetical protein POPTR_0008s17960g [Populus trichocarpa] |