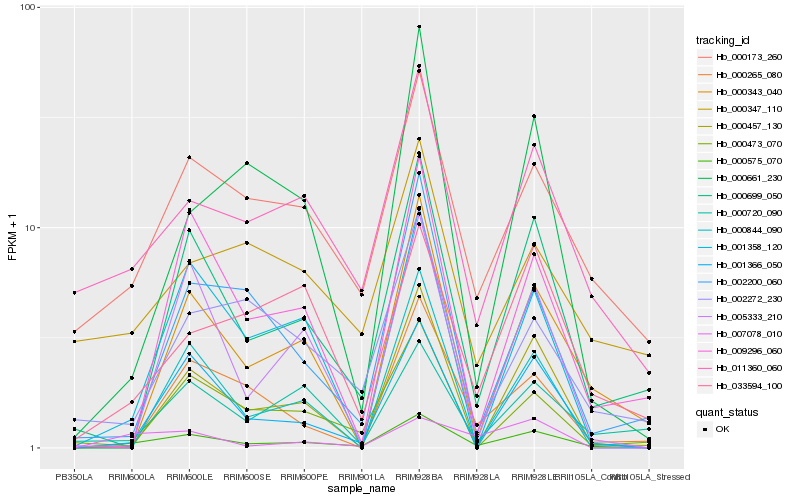
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000575_070 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X1 [Populus euphratica] |
| 2 |
Hb_000173_260 |
0.1855385301 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 3 |
Hb_011360_060 |
0.1869014995 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 4 |
Hb_001366_050 |
0.1951187138 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like isoform X2 [Populus euphratica] |
| 5 |
Hb_000699_050 |
0.2015252279 |
- |
- |
- |
| 6 |
Hb_001358_120 |
0.2278181738 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 7 |
Hb_000457_130 |
0.2281707422 |
- |
- |
hypothetical protein AALP_AA3G009100 [Arabis alpina] |
| 8 |
Hb_000265_080 |
0.2296327476 |
- |
- |
Vacuolar cation/proton exchanger, putative [Ricinus communis] |
| 9 |
Hb_000343_040 |
0.2303612382 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 10 |
Hb_005333_210 |
0.2329496415 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 11 |
Hb_000473_070 |
0.2335674926 |
- |
- |
PREDICTED: amino-acid permease BAT1 homolog [Jatropha curcas] |
| 12 |
Hb_033594_100 |
0.2349081897 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_000661_230 |
0.2355762221 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_007078_010 |
0.2399258962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002200_060 |
0.2400543151 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 16 |
Hb_000844_090 |
0.2421251614 |
- |
- |
PREDICTED: cytochrome P450 82C4-like [Jatropha curcas] |
| 17 |
Hb_002272_230 |
0.2432277231 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 18 |
Hb_009296_060 |
0.2446079296 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 19 |
Hb_000720_090 |
0.2453902774 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 20 |
Hb_000347_110 |
0.248128657 |
- |
- |
BnaA05g25330D [Brassica napus] |