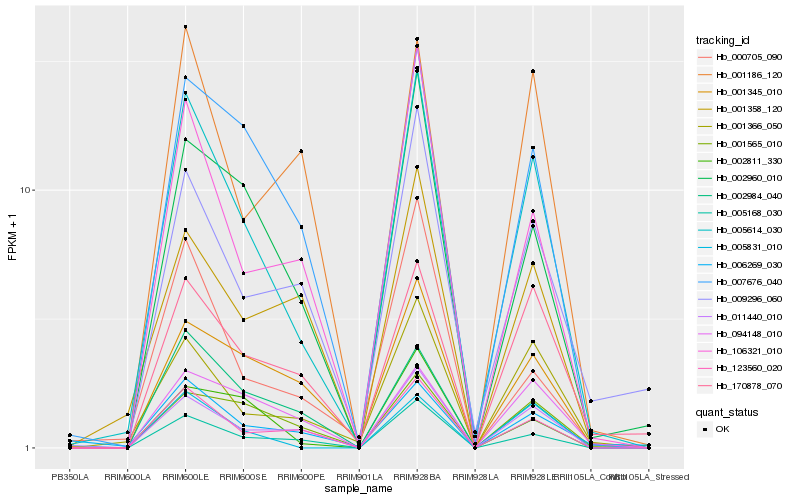
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005614_030 |
0.0 |
- |
- |
PREDICTED: transcription factor SCREAM2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_005168_030 |
0.1327421356 |
- |
- |
unknown [Zea mays] |
| 3 |
Hb_011440_010 |
0.1350456787 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 4 |
Hb_123560_020 |
0.1386965106 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |
| 5 |
Hb_001366_050 |
0.1389211871 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like isoform X2 [Populus euphratica] |
| 6 |
Hb_007676_040 |
0.1468468713 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 7 |
Hb_106321_010 |
0.1522262797 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 8 |
Hb_005831_010 |
0.1541703909 |
- |
- |
hypothetical protein JCGZ_04003 [Jatropha curcas] |
| 9 |
Hb_002960_010 |
0.1560270252 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 10 |
Hb_170878_070 |
0.1655173826 |
- |
- |
hypothetical protein EUGRSUZ_L008133, partial [Eucalyptus grandis] |
| 11 |
Hb_001345_010 |
0.1690907528 |
- |
- |
hypothetical protein MIMGU_mgv1a010889mg [Erythranthe guttata] |
| 12 |
Hb_009296_060 |
0.1713026422 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 13 |
Hb_002811_330 |
0.174238115 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 14 |
Hb_002984_040 |
0.1754226687 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_094148_010 |
0.1812838729 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_006269_030 |
0.1816873943 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 17 |
Hb_001358_120 |
0.182466144 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 18 |
Hb_001565_010 |
0.18342944 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 19 |
Hb_000705_090 |
0.1852106251 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 20 |
Hb_001186_120 |
0.1858256676 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |