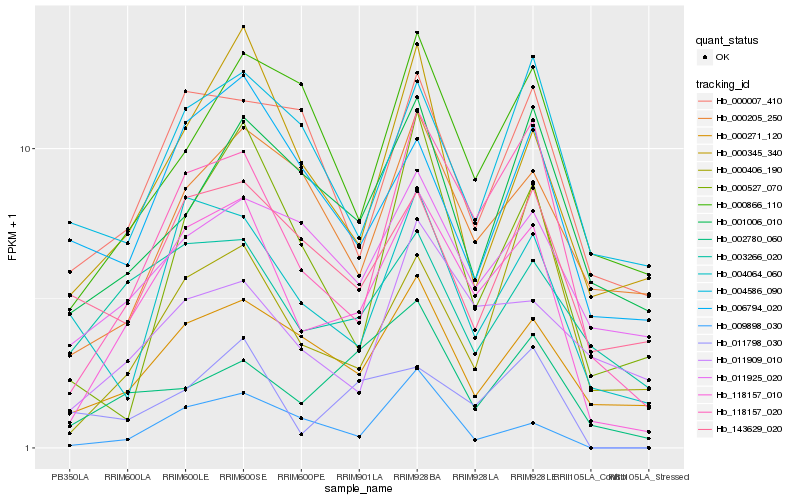
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_118157_010 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 2 |
Hb_118157_020 |
0.1150735456 |
- |
- |
hypothetical protein MTR_3g456110 [Medicago truncatula] |
| 3 |
Hb_000271_120 |
0.1799925309 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 4 |
Hb_000345_340 |
0.1907807389 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 5 |
Hb_004064_060 |
0.1991029245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003266_020 |
0.2006896201 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 7 |
Hb_000007_410 |
0.2016265249 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 8 |
Hb_000866_110 |
0.2021592152 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002780_060 |
0.204089296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011925_020 |
0.2053094797 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_000406_190 |
0.2059633301 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 12 |
Hb_000205_250 |
0.2071669707 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 13 |
Hb_011909_010 |
0.208622001 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 14 |
Hb_006794_020 |
0.2095733306 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 15 |
Hb_001006_010 |
0.2103356076 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 16 |
Hb_000527_070 |
0.2106819729 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 17 |
Hb_011798_030 |
0.2124116319 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Vitis vinifera] |
| 18 |
Hb_143629_020 |
0.2125684423 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 19 |
Hb_004586_090 |
0.2162798244 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009898_030 |
0.2166471525 |
- |
- |
PREDICTED: uncharacterized protein LOC103720281 [Phoenix dactylifera] |