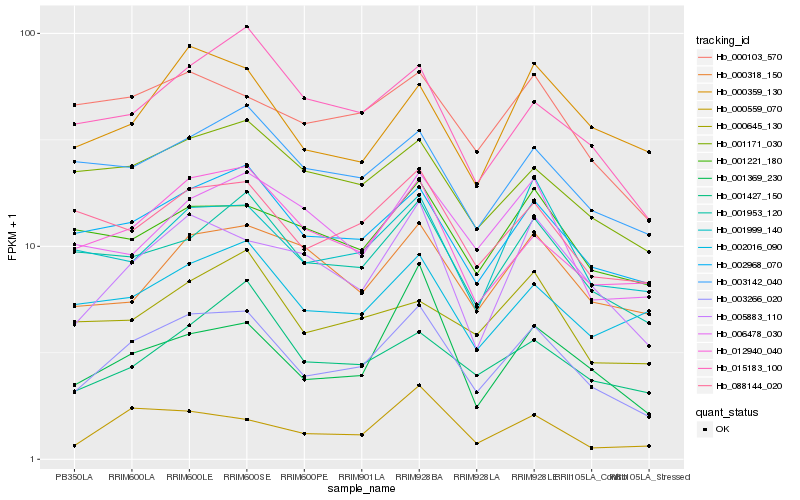
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003266_020 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 2 |
Hb_002968_070 |
0.0993292104 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 3 |
Hb_005883_110 |
0.1005664712 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001953_120 |
0.1108844466 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 5 |
Hb_012940_040 |
0.1116056501 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 6 |
Hb_015183_100 |
0.1191371946 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 7 |
Hb_088144_020 |
0.1202867641 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001171_030 |
0.1219210088 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 9 |
Hb_000559_070 |
0.1224794335 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 10 |
Hb_003142_040 |
0.1227249251 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 11 |
Hb_001999_140 |
0.1234083798 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_000103_570 |
0.1250266993 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000359_130 |
0.1259505588 |
- |
- |
4-hydroxyphenylpyruvate dioxygenase [Hevea brasiliensis] |
| 14 |
Hb_001369_230 |
0.1264881378 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 15 |
Hb_002016_090 |
0.1287228915 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 16 |
Hb_001427_150 |
0.1300374485 |
- |
- |
PREDICTED: uncharacterized protein LOC105637790 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006478_030 |
0.1300773859 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 18 |
Hb_000318_150 |
0.1303746022 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 19 |
Hb_001221_180 |
0.1304652516 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 20 |
Hb_000645_130 |
0.1308300917 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |