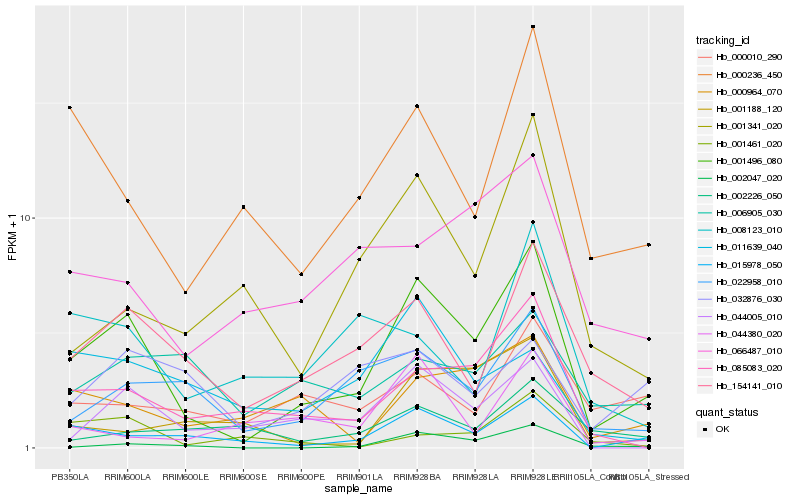
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001496_080 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 2 |
Hb_015978_050 |
0.2142586536 |
- |
- |
- |
| 3 |
Hb_154141_010 |
0.2253323746 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 4 |
Hb_002047_020 |
0.2342618 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_085083_020 |
0.237222613 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 6 |
Hb_022958_010 |
0.2415448005 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 7 |
Hb_000236_450 |
0.2457249346 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 8 |
Hb_044005_010 |
0.2510813871 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 9 |
Hb_000964_070 |
0.259276006 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 10 |
Hb_001341_020 |
0.2634100319 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 11 |
Hb_066487_010 |
0.2702340597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000010_290 |
0.2724557042 |
- |
- |
- |
| 13 |
Hb_006905_030 |
0.2844204652 |
- |
- |
- |
| 14 |
Hb_032876_030 |
0.2889080822 |
- |
- |
- |
| 15 |
Hb_008123_010 |
0.2920260871 |
- |
- |
- |
| 16 |
Hb_001461_020 |
0.2931665039 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_002226_050 |
0.2935653772 |
- |
- |
PREDICTED: uncharacterized protein LOC105641366 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001188_120 |
0.29440772 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 19 |
Hb_044380_020 |
0.2945414683 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 20 |
Hb_011639_040 |
0.2945498096 |
- |
- |
hypothetical protein M569_08265, partial [Genlisea aurea] |