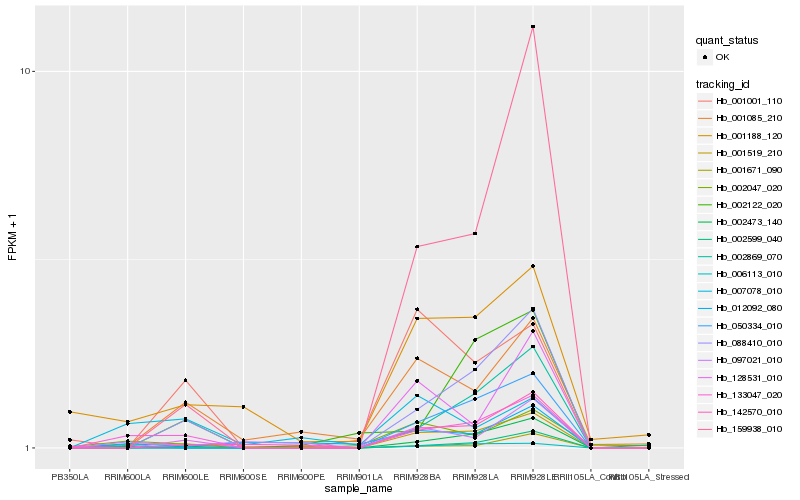
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_133047_020 |
0.0 |
- |
- |
lysophosphatidic acid acyltransferase, putative [Ricinus communis] |
| 2 |
Hb_088410_010 |
0.2329987607 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_002473_140 |
0.2632614132 |
- |
- |
- |
| 4 |
Hb_006113_010 |
0.2703766639 |
- |
- |
hypothetical protein VITISV_015014 [Vitis vinifera] |
| 5 |
Hb_002047_020 |
0.2778229094 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001671_090 |
0.2837574836 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_159938_010 |
0.297792746 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 8 |
Hb_001188_120 |
0.30544832 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 9 |
Hb_097021_010 |
0.3068791217 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 10 |
Hb_007078_010 |
0.3082457866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_142570_010 |
0.3091841977 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_001085_210 |
0.3148907556 |
- |
- |
PREDICTED: myosin-binding protein 3 [Jatropha curcas] |
| 13 |
Hb_002122_020 |
0.3225641747 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 14 |
Hb_001519_210 |
0.3246573701 |
- |
- |
hypothetical protein B456_003G032500 [Gossypium raimondii] |
| 15 |
Hb_050334_010 |
0.3253189001 |
- |
- |
- |
| 16 |
Hb_012092_080 |
0.3301053124 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280-like [Malus domestica] |
| 17 |
Hb_128531_010 |
0.3329882349 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 18 |
Hb_001001_110 |
0.3339528607 |
- |
- |
- |
| 19 |
Hb_002869_070 |
0.3347841104 |
- |
- |
- |
| 20 |
Hb_002599_040 |
0.3372697247 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |