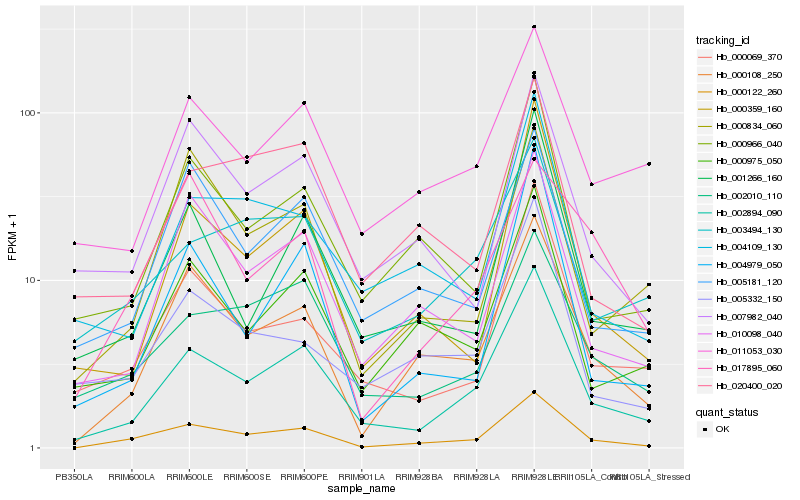
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000122_260 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 1-like [Jatropha curcas] |
| 2 |
Hb_000108_250 |
0.1141549216 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002894_090 |
0.1308901012 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase [Jatropha curcas] |
| 4 |
Hb_002010_110 |
0.1737823196 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 5 |
Hb_010098_040 |
0.1741610478 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 6 |
Hb_000975_050 |
0.1769797575 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000966_040 |
0.1806133725 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 8 |
Hb_005181_120 |
0.1833012593 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_007982_040 |
0.1843420028 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003494_130 |
0.1847469097 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 11 |
Hb_000069_370 |
0.185397392 |
- |
- |
- |
| 12 |
Hb_000359_160 |
0.1903034088 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 13 |
Hb_005332_150 |
0.1931068447 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |
| 14 |
Hb_011053_030 |
0.1943394562 |
- |
- |
- |
| 15 |
Hb_017895_060 |
0.1946663955 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 14, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001266_160 |
0.1969578514 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_004109_130 |
0.1978129553 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000834_060 |
0.1979598645 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_020400_020 |
0.1983980582 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 20 |
Hb_004979_050 |
0.2007897072 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |