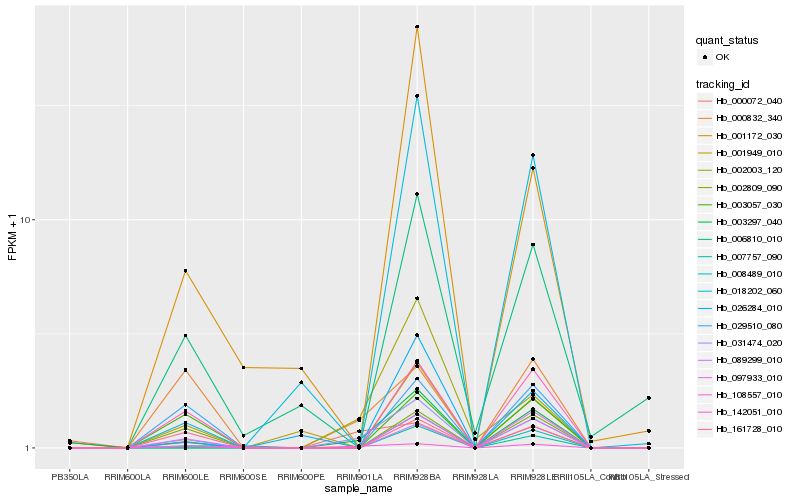
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031474_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002003_120 |
0.2450557069 |
- |
- |
PREDICTED: ubiquitin-like-conjugating enzyme ATG10 isoform X1 [Jatropha curcas] |
| 3 |
Hb_142051_010 |
0.2450898404 |
- |
- |
- |
| 4 |
Hb_003297_040 |
0.2469935675 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 5 |
Hb_108557_010 |
0.2573746949 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 6 |
Hb_008489_010 |
0.2623560409 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 7 |
Hb_000832_340 |
0.2701323293 |
- |
- |
PREDICTED: psbP-like protein 2, chloroplastic isoform X3 [Nelumbo nucifera] |
| 8 |
Hb_002809_090 |
0.2722699193 |
- |
- |
- |
| 9 |
Hb_161728_010 |
0.2741810737 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 10 |
Hb_089299_010 |
0.2761433581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_097933_010 |
0.2800894206 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_006810_010 |
0.2827976896 |
- |
- |
hypothetical protein B456_012G018200 [Gossypium raimondii] |
| 13 |
Hb_003057_030 |
0.2831578905 |
- |
- |
hypothetical protein JCGZ_06668 [Jatropha curcas] |
| 14 |
Hb_029510_080 |
0.283241186 |
- |
- |
- |
| 15 |
Hb_000072_040 |
0.2854170453 |
- |
- |
- |
| 16 |
Hb_001172_030 |
0.2859950124 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 7.2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_018202_060 |
0.286803045 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Jatropha curcas] |
| 18 |
Hb_001949_010 |
0.2890344206 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 19 |
Hb_026284_010 |
0.2904707254 |
- |
- |
- |
| 20 |
Hb_007757_090 |
0.2933661637 |
- |
- |
PREDICTED: uncharacterized protein LOC105793478 [Gossypium raimondii] |