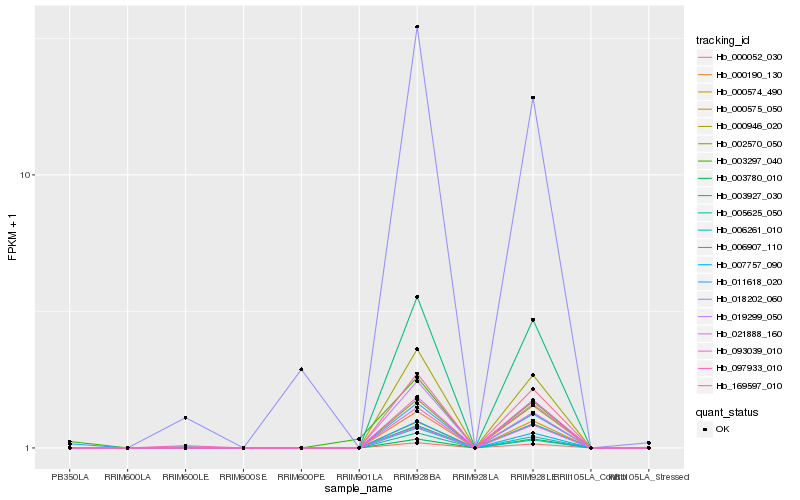
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011618_020 |
0.0 |
- |
- |
hypothetical protein OsI_13615 [Oryza sativa Indica Group] |
| 2 |
Hb_169597_010 |
0.1653825464 |
- |
- |
PREDICTED: uncharacterized protein LOC104903465 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_003927_030 |
0.1655999519 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_000052_030 |
0.1656815536 |
- |
- |
- |
| 5 |
Hb_000946_020 |
0.1665616731 |
- |
- |
hypothetical protein PRUPE_ppa023262mg [Prunus persica] |
| 6 |
Hb_002570_050 |
0.1687554823 |
- |
- |
PREDICTED: uncharacterized protein LOC104897677 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_093039_010 |
0.1732967994 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 8 |
Hb_097933_010 |
0.1737912728 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_006261_010 |
0.1741375318 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 10 |
Hb_000190_130 |
0.1744132473 |
- |
- |
cysteine protease inhibitor [Populus tremula] |
| 11 |
Hb_007757_090 |
0.1762244403 |
- |
- |
PREDICTED: uncharacterized protein LOC105793478 [Gossypium raimondii] |
| 12 |
Hb_005625_050 |
0.1790281815 |
- |
- |
hypothetical protein PRUPE_ppa024499mg, partial [Prunus persica] |
| 13 |
Hb_003780_010 |
0.1802696517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000574_490 |
0.182574352 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 11 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006907_110 |
0.1859738735 |
- |
- |
- |
| 16 |
Hb_000575_050 |
0.1863809282 |
- |
- |
hypothetical protein CISIN_1g0468601mg, partial [Citrus sinensis] |
| 17 |
Hb_018202_060 |
0.1868238552 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Jatropha curcas] |
| 18 |
Hb_003297_040 |
0.1870754938 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 19 |
Hb_021888_160 |
0.1874606808 |
- |
- |
- |
| 20 |
Hb_019299_050 |
0.1883210043 |
- |
- |
- |