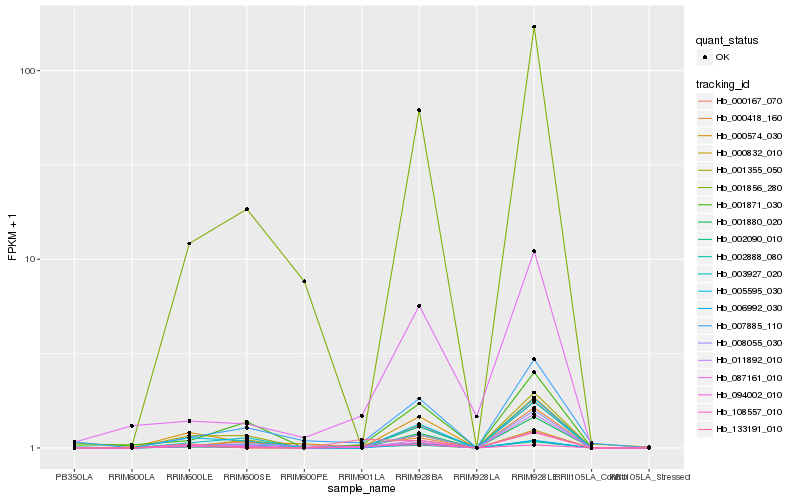
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002090_010 |
0.0 |
- |
- |
gag-pol fusion protein [Rhizophagus irregularis DAOM 197198w] |
| 2 |
Hb_094002_010 |
0.123714701 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 3 |
Hb_008055_030 |
0.2100834373 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 4 |
Hb_005595_030 |
0.2124487606 |
- |
- |
hypothetical protein EUGRSUZ_A00624 [Eucalyptus grandis] |
| 5 |
Hb_001880_020 |
0.215790846 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 6 |
Hb_108557_010 |
0.2160609292 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 7 |
Hb_000418_160 |
0.219347221 |
- |
- |
- |
| 8 |
Hb_003927_020 |
0.2292363004 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |
| 9 |
Hb_000167_070 |
0.2329396728 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 10 |
Hb_001355_050 |
0.2340434385 |
- |
- |
- |
| 11 |
Hb_087161_010 |
0.2345896605 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_007885_110 |
0.2355378686 |
- |
- |
PREDICTED: uncharacterized protein LOC104109376 [Nicotiana tomentosiformis] |
| 13 |
Hb_001871_030 |
0.2362331487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002888_080 |
0.2362363119 |
- |
- |
PREDICTED: uncharacterized protein LOC104120414 [Nicotiana tomentosiformis] |
| 15 |
Hb_000574_030 |
0.2412331706 |
desease resistance |
Gene Name: Glyco_hydro_38 |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 16 |
Hb_011892_010 |
0.2420575743 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_133191_010 |
0.2426123297 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 18 |
Hb_006992_030 |
0.2449310676 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_001856_280 |
0.2474605288 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 20 |
Hb_000832_010 |
0.2505755672 |
- |
- |
hypothetical protein VITISV_005044 [Vitis vinifera] |