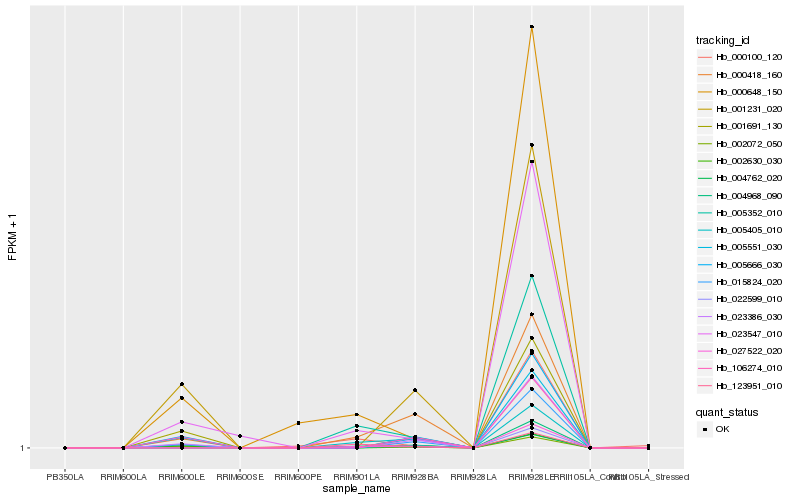
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002072_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_000418_160 |
0.1440110581 |
- |
- |
- |
| 3 |
Hb_023547_010 |
0.2021090055 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_005352_010 |
0.2086964967 |
- |
- |
- |
| 5 |
Hb_022599_010 |
0.209006786 |
- |
- |
Uncharacterized protein TCM_010507 [Theobroma cacao] |
| 6 |
Hb_004762_020 |
0.2141136264 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_123951_010 |
0.2176503205 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_005405_010 |
0.2179048523 |
- |
- |
- |
| 9 |
Hb_004968_090 |
0.2182564083 |
- |
- |
PREDICTED: uncharacterized protein LOC104882781 [Vitis vinifera] |
| 10 |
Hb_005666_030 |
0.2183497748 |
- |
- |
PREDICTED: uncharacterized protein LOC103489421 [Cucumis melo] |
| 11 |
Hb_027522_020 |
0.2193675317 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_005551_030 |
0.2195586525 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 13 |
Hb_000648_150 |
0.2212472693 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase V.1 [Camelina sativa] |
| 14 |
Hb_001691_130 |
0.2214511226 |
- |
- |
PREDICTED: uncharacterized protein LOC104587431 [Nelumbo nucifera] |
| 15 |
Hb_001231_020 |
0.2217669592 |
- |
- |
- |
| 16 |
Hb_000100_120 |
0.2227967358 |
- |
- |
RNA-directed DNA polymerase (Reverse transcriptase); Chromo; Zinc finger, CCHC-type; Peptidase aspartic, active site; Polynucleotidyl transferase, Ribonuclease H fold [Medicago truncatula] |
| 17 |
Hb_002630_030 |
0.2251242692 |
- |
- |
OSIGBa0132I10.1 [Oryza sativa Indica Group] |
| 18 |
Hb_106274_010 |
0.2254217509 |
- |
- |
hypothetical protein VITISV_008825 [Vitis vinifera] |
| 19 |
Hb_015824_020 |
0.2264251473 |
- |
- |
- |
| 20 |
Hb_023386_030 |
0.2274976886 |
- |
- |
- |