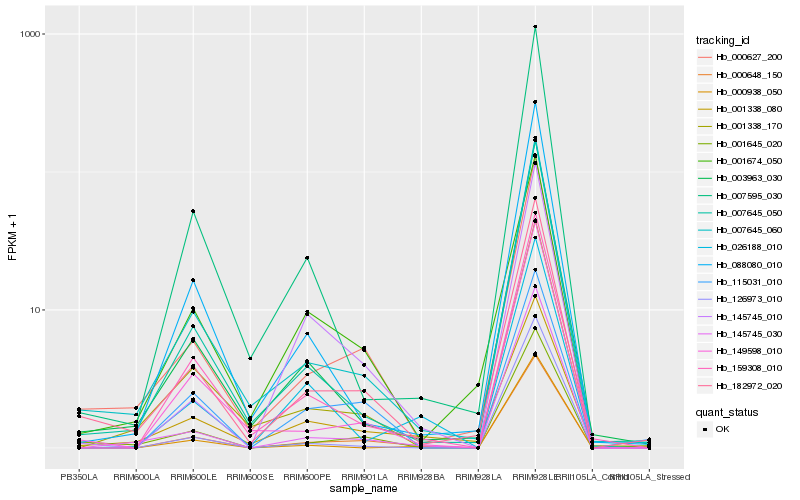
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000648_150 |
0.0 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase V.1 [Camelina sativa] |
| 2 |
Hb_159308_010 |
0.1015505205 |
- |
- |
ribosomal protein S4 [Cercidiphyllum japonicum] |
| 3 |
Hb_000627_200 |
0.1129610686 |
- |
- |
NADH dehydrogenase subunit I [Hevea brasiliensis] |
| 4 |
Hb_115031_010 |
0.1135494475 |
- |
- |
photosystem II P680 chlorophyll A apoprotein, partial (chloroplast) [Oenothera grandiflora] |
| 5 |
Hb_007645_060 |
0.1139808905 |
- |
- |
cytochrome f [Hevea brasiliensis] |
| 6 |
Hb_182972_020 |
0.1154495517 |
- |
- |
cytochrome b6 [Gossypium barbadense] |
| 7 |
Hb_145745_030 |
0.1184398968 |
- |
- |
BnaC09g26890D [Brassica napus] |
| 8 |
Hb_149598_010 |
0.1190789628 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |
| 9 |
Hb_001338_170 |
0.1201662901 |
- |
- |
NADH dehydrogenase subunit 4 [Hevea brasiliensis] |
| 10 |
Hb_007645_050 |
0.1279908499 |
- |
- |
acetyl-CoA carboxylase beta subunit [Hevea brasiliensis] |
| 11 |
Hb_088080_010 |
0.1324715471 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 12 |
Hb_001338_080 |
0.1329149754 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Tamarindus indica] |
| 13 |
Hb_026188_010 |
0.1341700063 |
- |
- |
PREDICTED: tropinone reductase homolog [Jatropha curcas] |
| 14 |
Hb_007595_030 |
0.1344777255 |
- |
- |
psbA gene product (chloroplast) [Silene noctiflora] |
| 15 |
Hb_000938_050 |
0.1365663913 |
- |
- |
hypothetical protein JCGZ_09640 [Jatropha curcas] |
| 16 |
Hb_001645_020 |
0.1370864934 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit H [Medicago truncatula] |
| 17 |
Hb_145745_010 |
0.1385273951 |
- |
- |
BnaUnng00830D [Brassica napus] |
| 18 |
Hb_001674_050 |
0.138854442 |
- |
- |
cytochrome b6 (chloroplast) [Apios americana] |
| 19 |
Hb_003963_030 |
0.1406434241 |
- |
- |
psbB [Jatropha curcas] |
| 20 |
Hb_126973_010 |
0.141082834 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Vitis vinifera] |