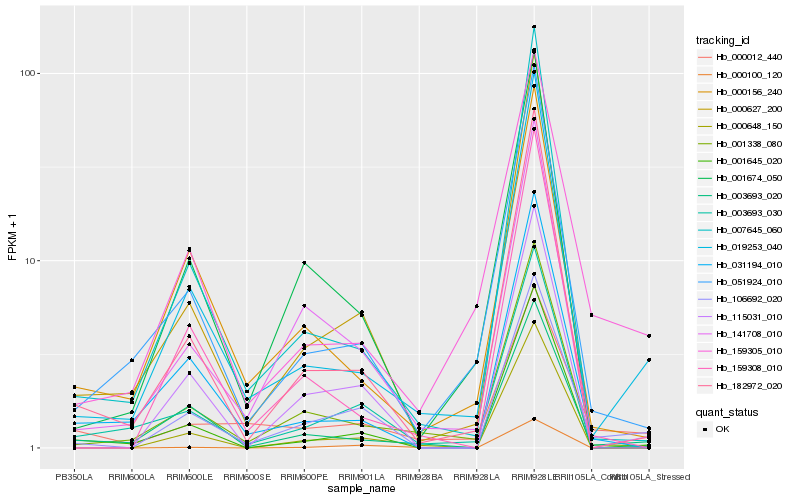
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106692_020 |
0.0 |
- |
- |
NADH dehydrogenase subunit F, partial (chloroplast) [Hevea sp. Gillespie 4272] |
| 2 |
Hb_115031_010 |
0.1318889146 |
- |
- |
photosystem II P680 chlorophyll A apoprotein, partial (chloroplast) [Oenothera grandiflora] |
| 3 |
Hb_019253_040 |
0.1753021758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003693_020 |
0.178340203 |
- |
- |
Ycf1, partial (chloroplast) [Clusia rosea] |
| 5 |
Hb_001645_020 |
0.1790183811 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit H [Medicago truncatula] |
| 6 |
Hb_182972_020 |
0.1813499651 |
- |
- |
cytochrome b6 [Gossypium barbadense] |
| 7 |
Hb_003693_030 |
0.1845120144 |
- |
- |
NADH dehydrogenase subunit 7 [Hevea brasiliensis] |
| 8 |
Hb_141708_010 |
0.1866498644 |
- |
- |
hypothetical protein EUTSA_v10011731mg [Eutrema salsugineum] |
| 9 |
Hb_000100_120 |
0.1868153447 |
- |
- |
RNA-directed DNA polymerase (Reverse transcriptase); Chromo; Zinc finger, CCHC-type; Peptidase aspartic, active site; Polynucleotidyl transferase, Ribonuclease H fold [Medicago truncatula] |
| 10 |
Hb_001338_080 |
0.1877914849 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Tamarindus indica] |
| 11 |
Hb_001674_050 |
0.1899642754 |
- |
- |
cytochrome b6 (chloroplast) [Apios americana] |
| 12 |
Hb_000648_150 |
0.1903397909 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase V.1 [Camelina sativa] |
| 13 |
Hb_000627_200 |
0.1903580257 |
- |
- |
NADH dehydrogenase subunit I [Hevea brasiliensis] |
| 14 |
Hb_159305_010 |
0.1905988838 |
- |
- |
rpl2 [Jatropha curcas] |
| 15 |
Hb_051924_010 |
0.1967614885 |
- |
- |
BnaA01g34100D [Brassica napus] |
| 16 |
Hb_000012_440 |
0.1974504542 |
- |
- |
PREDICTED: uncharacterized protein LOC104248335 [Nicotiana sylvestris] |
| 17 |
Hb_031194_010 |
0.1988502229 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Arachis hypogaea] |
| 18 |
Hb_000156_240 |
0.2010945638 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 19 |
Hb_007645_060 |
0.2013373344 |
- |
- |
cytochrome f [Hevea brasiliensis] |
| 20 |
Hb_159308_010 |
0.2020275959 |
- |
- |
ribosomal protein S4 [Cercidiphyllum japonicum] |