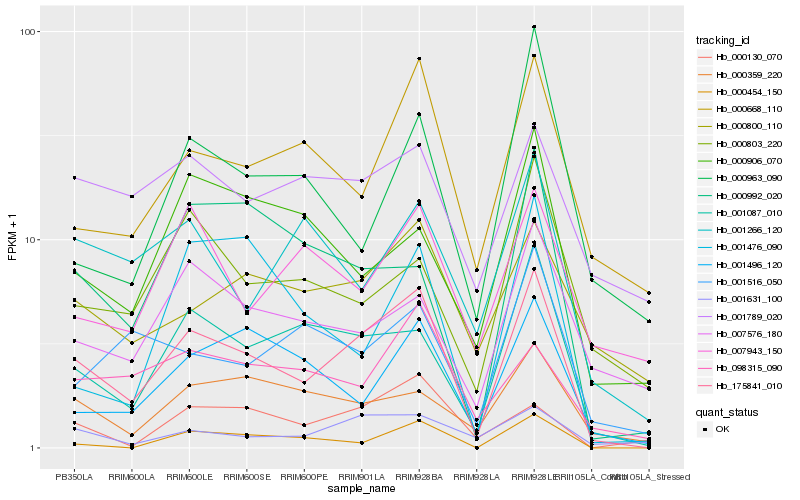
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_175841_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 2 |
Hb_001087_010 |
0.1793660115 |
- |
- |
PREDICTED: uncharacterized protein LOC105630042 [Jatropha curcas] |
| 3 |
Hb_000668_110 |
0.2050115534 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 4 |
Hb_001631_100 |
0.2118707858 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001496_120 |
0.2148411115 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_000800_110 |
0.2180868181 |
- |
- |
NAD dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000454_150 |
0.2195038479 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 8 |
Hb_000359_220 |
0.2197154587 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 9 |
Hb_098315_090 |
0.2213571919 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 10 |
Hb_007576_180 |
0.2244515882 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 11 |
Hb_001266_120 |
0.2247625011 |
- |
- |
auxilin, putative [Ricinus communis] |
| 12 |
Hb_000130_070 |
0.2254866946 |
- |
- |
hypothetical protein JCGZ_14738 [Jatropha curcas] |
| 13 |
Hb_000803_220 |
0.2278401182 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001516_050 |
0.2292356899 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 15 |
Hb_000992_020 |
0.2298278037 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 16 |
Hb_001789_020 |
0.2342067973 |
- |
- |
PREDICTED: tripeptidyl-peptidase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007943_150 |
0.2351645418 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_000906_070 |
0.2369827071 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000963_090 |
0.2386989168 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_001476_090 |
0.238893001 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |