| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
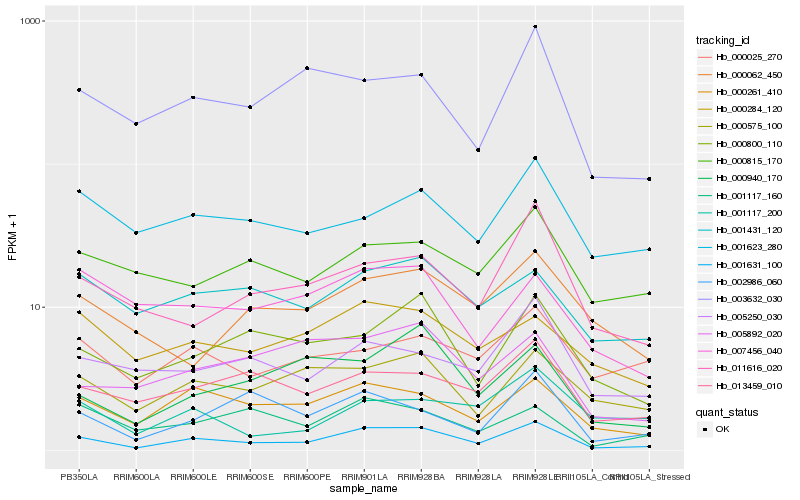
Hb_001631_100 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000261_410 |
0.1584711698 |
- |
- |
PREDICTED: myosin-binding protein 3 [Jatropha curcas] |
| 3 |
Hb_000800_110 |
0.1731895756 |
- |
- |
NAD dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_000575_100 |
0.1754052638 |
- |
- |
PREDICTED: nephrocystin-3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_013459_010 |
0.1768363373 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 6 |
Hb_011616_020 |
0.1786040304 |
- |
- |
hypothetical protein CICLE_v10004968mg [Citrus clementina] |
| 7 |
Hb_003632_030 |
0.1793934181 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 8 |
Hb_000940_170 |
0.1851406363 |
- |
- |
hypothetical protein VITISV_029834 [Vitis vinifera] |
| 9 |
Hb_005250_030 |
0.1880949902 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 10 |
Hb_002986_060 |
0.1884110711 |
- |
- |
hypothetical protein M569_15879, partial [Genlisea aurea] |
| 11 |
Hb_001431_120 |
0.1897194104 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 12 |
Hb_000284_120 |
0.1902907769 |
- |
- |
PREDICTED: uncharacterized protein LOC105629656 [Jatropha curcas] |
| 13 |
Hb_005892_020 |
0.1952725027 |
- |
- |
PREDICTED: uncharacterized protein LOC105632170 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001117_160 |
0.1956382173 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 15 |
Hb_001623_280 |
0.1959960247 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000815_170 |
0.1961467861 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 17 |
Hb_000025_270 |
0.1975314818 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_001117_200 |
0.2003295661 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000062_450 |
0.2006098593 |
- |
- |
hypothetical protein MTR_081s0009, partial [Medicago truncatula] |
| 20 |
Hb_007456_040 |
0.2012268428 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |