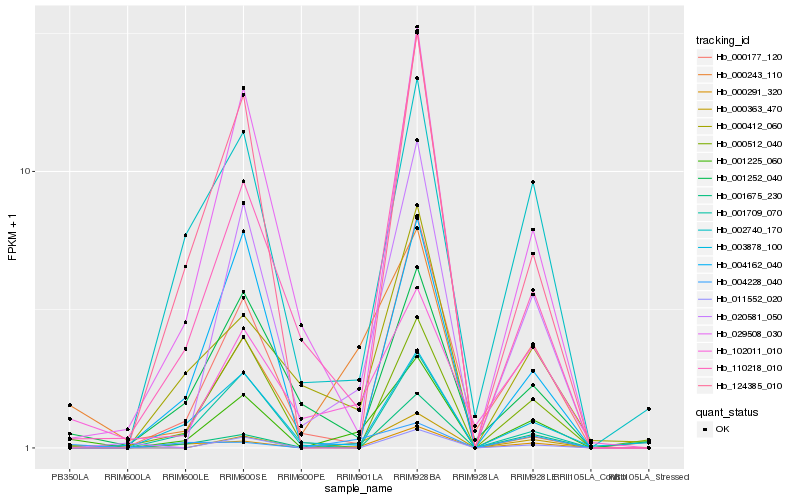
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001225_060 |
0.0 |
- |
- |
BnaC03g58850D [Brassica napus] |
| 2 |
Hb_000363_470 |
0.1423389513 |
- |
- |
hypothetical protein JCGZ_00950 [Jatropha curcas] |
| 3 |
Hb_000291_320 |
0.2054524627 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 4 |
Hb_020581_050 |
0.2085754409 |
- |
- |
- |
| 5 |
Hb_102011_010 |
0.2327743099 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 6 |
Hb_000177_120 |
0.2384379832 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004228_040 |
0.2466213341 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 8 |
Hb_029508_030 |
0.2472295771 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0002s04880g [Populus trichocarpa] |
| 9 |
Hb_124385_010 |
0.2481167844 |
- |
- |
hypothetical protein JCGZ_13882 [Jatropha curcas] |
| 10 |
Hb_001675_230 |
0.2500111466 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 11 |
Hb_004162_040 |
0.2512632739 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase-like [Crassostrea gigas] |
| 12 |
Hb_002740_170 |
0.2536665886 |
- |
- |
PREDICTED: uncharacterized protein LOC105647357 [Jatropha curcas] |
| 13 |
Hb_000512_040 |
0.2560106529 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_001252_040 |
0.2565170705 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 15 |
Hb_110218_010 |
0.2594409967 |
- |
- |
PREDICTED: uncharacterized protein LOC104428128 [Eucalyptus grandis] |
| 16 |
Hb_000412_060 |
0.260998581 |
- |
- |
hypothetical protein JCGZ_18814 [Jatropha curcas] |
| 17 |
Hb_000243_110 |
0.2616219623 |
- |
- |
PREDICTED: WAT1-related protein At3g53210-like [Jatropha curcas] |
| 18 |
Hb_003878_100 |
0.2628898843 |
desease resistance |
Gene Name: NB-ARC |
LRR and NB-ARC domains-containing disease resistance protein, putative [Theobroma cacao] |
| 19 |
Hb_011552_020 |
0.2650767479 |
- |
- |
hypothetical protein F383_28301 [Gossypium arboreum] |
| 20 |
Hb_001709_070 |
0.2659058467 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |