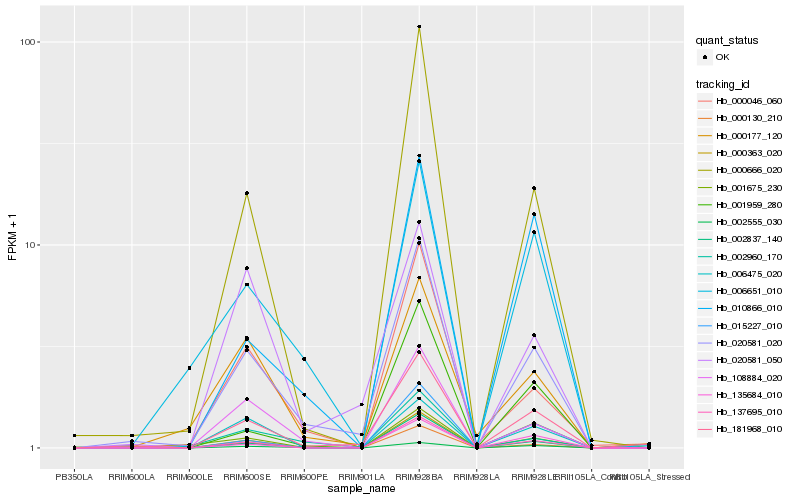
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_181968_010 |
0.0 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 2 |
Hb_020581_020 |
0.1137867318 |
- |
- |
hypothetical protein B456_003G065300 [Gossypium raimondii] |
| 3 |
Hb_137695_010 |
0.1560608193 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Malus domestica] |
| 4 |
Hb_002837_140 |
0.1659454513 |
- |
- |
hypothetical protein JCGZ_05022 [Jatropha curcas] |
| 5 |
Hb_000666_020 |
0.1660090565 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 6 |
Hb_000130_210 |
0.1751988147 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Populus euphratica] |
| 7 |
Hb_135684_010 |
0.1782066428 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |
| 8 |
Hb_015227_010 |
0.1802657448 |
- |
- |
PREDICTED: uncharacterized protein LOC104903465 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_000046_060 |
0.1924506921 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 10 |
Hb_002555_030 |
0.1971645362 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Malus domestica] |
| 11 |
Hb_001959_280 |
0.1991508381 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 12 |
Hb_108884_020 |
0.2044755946 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 13 |
Hb_010866_010 |
0.2073111871 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 14 |
Hb_001675_230 |
0.2090921357 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 15 |
Hb_002960_170 |
0.2131591894 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 16 |
Hb_000177_120 |
0.2134826082 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006651_010 |
0.2158993966 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4-like [Citrus sinensis] |
| 18 |
Hb_020581_050 |
0.2160162856 |
- |
- |
- |
| 19 |
Hb_000363_020 |
0.2166392011 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_006475_020 |
0.2239332108 |
- |
- |
PREDICTED: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |