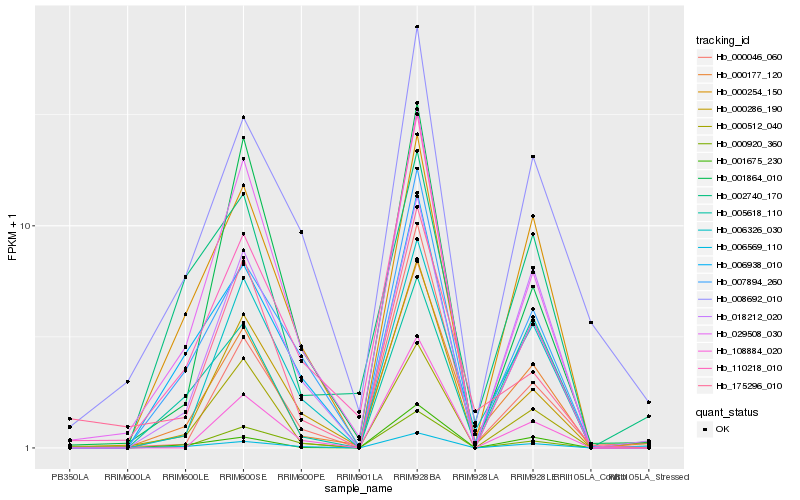
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000177_120 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000286_190 |
0.1388639815 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 3 |
Hb_006326_030 |
0.1398647192 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_029508_030 |
0.143156714 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0002s04880g [Populus trichocarpa] |
| 5 |
Hb_007894_260 |
0.1437155586 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_006569_110 |
0.1500318458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000254_150 |
0.1550125614 |
- |
- |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 8 |
Hb_018212_020 |
0.1559940737 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 9 |
Hb_000512_040 |
0.162127305 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 10 |
Hb_006938_010 |
0.1661060804 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 11 |
Hb_001675_230 |
0.1688147218 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 12 |
Hb_008692_010 |
0.1690545066 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 13 |
Hb_000920_360 |
0.1705901422 |
- |
- |
PREDICTED: (R,S)-reticuline 7-O-methyltransferase-like [Jatropha curcas] |
| 14 |
Hb_000046_060 |
0.1725785865 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 15 |
Hb_175296_010 |
0.1727672661 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 16 |
Hb_108884_020 |
0.1736482773 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 17 |
Hb_110218_010 |
0.1753423059 |
- |
- |
PREDICTED: uncharacterized protein LOC104428128 [Eucalyptus grandis] |
| 18 |
Hb_001864_010 |
0.1777406474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005618_110 |
0.1810113904 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 20 |
Hb_002740_170 |
0.1814892222 |
- |
- |
PREDICTED: uncharacterized protein LOC105647357 [Jatropha curcas] |