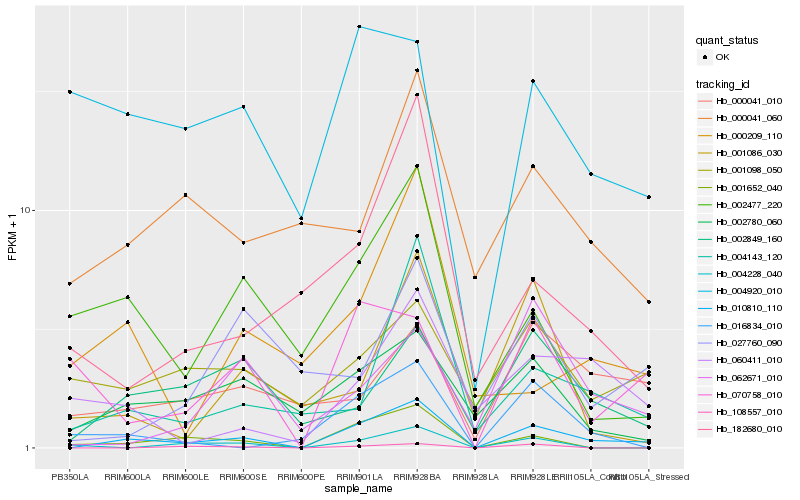
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010810_110 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 2 |
Hb_001652_040 |
0.280554215 |
- |
- |
PREDICTED: uncharacterized protein LOC103341683 [Prunus mume] |
| 3 |
Hb_004143_120 |
0.3011116276 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 4 |
Hb_002780_060 |
0.3035468103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002849_160 |
0.3052224644 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 6 |
Hb_016834_010 |
0.315462467 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 7 |
Hb_002477_220 |
0.3180071189 |
- |
- |
hypothetical protein POPTR_0017s06650g [Populus trichocarpa] |
| 8 |
Hb_182680_010 |
0.3190798526 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 9 |
Hb_060411_010 |
0.3208363311 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 10 |
Hb_070758_010 |
0.3227978471 |
- |
- |
- |
| 11 |
Hb_000041_010 |
0.3300565479 |
- |
- |
pfkB-type carbohydrate kinase family protein [Populus trichocarpa] |
| 12 |
Hb_000041_060 |
0.3304318226 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_001098_050 |
0.3310858191 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 14 |
Hb_062671_010 |
0.3318178093 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004228_040 |
0.3381341667 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 16 |
Hb_027760_090 |
0.3383285875 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_004920_010 |
0.3387876011 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |
| 18 |
Hb_108557_010 |
0.3389342793 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 19 |
Hb_000209_110 |
0.3431593419 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 20 |
Hb_001086_030 |
0.3437294332 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |