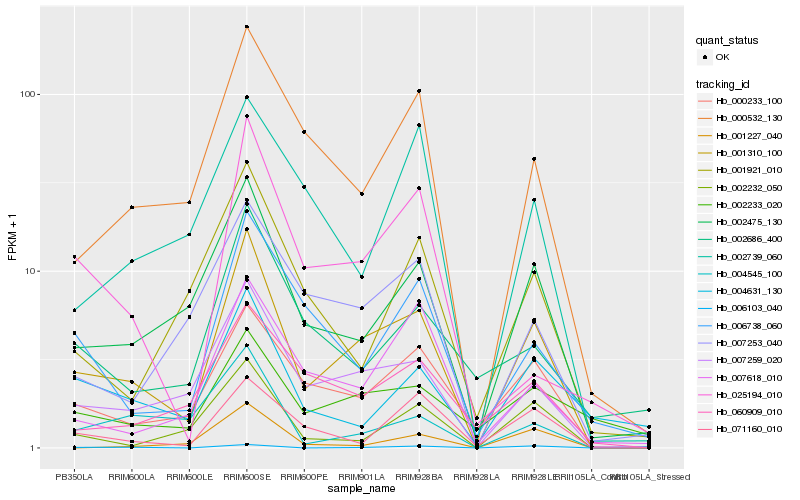
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001310_100 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable N-acetyltransferase HLS1 [Jatropha curcas] |
| 2 |
Hb_007259_020 |
0.1547487691 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 3 |
Hb_002475_130 |
0.1785553562 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 4 [Jatropha curcas] |
| 4 |
Hb_000233_100 |
0.1880486401 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 5 |
Hb_000532_130 |
0.2033425737 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 6 |
Hb_002232_050 |
0.2144303342 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 7 |
Hb_006103_040 |
0.2246213471 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_006738_060 |
0.2273820667 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_007253_040 |
0.2304909812 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 10 |
Hb_060909_010 |
0.2323814753 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 11 |
Hb_007618_010 |
0.2355116792 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 12 |
Hb_071160_010 |
0.2374209326 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 13 |
Hb_004631_130 |
0.2378785476 |
- |
- |
helicase domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_004545_100 |
0.2398261466 |
- |
- |
PREDICTED: protein BONZAI 3 [Jatropha curcas] |
| 15 |
Hb_002233_020 |
0.2414680417 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 16 |
Hb_025194_010 |
0.2461748397 |
- |
- |
PREDICTED: uncharacterized protein LOC105644857 [Jatropha curcas] |
| 17 |
Hb_001921_010 |
0.2467796588 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 18 |
Hb_002686_400 |
0.2490394294 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 19 |
Hb_001227_040 |
0.2495458166 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 20 |
Hb_002739_060 |
0.2517732856 |
- |
- |
- |