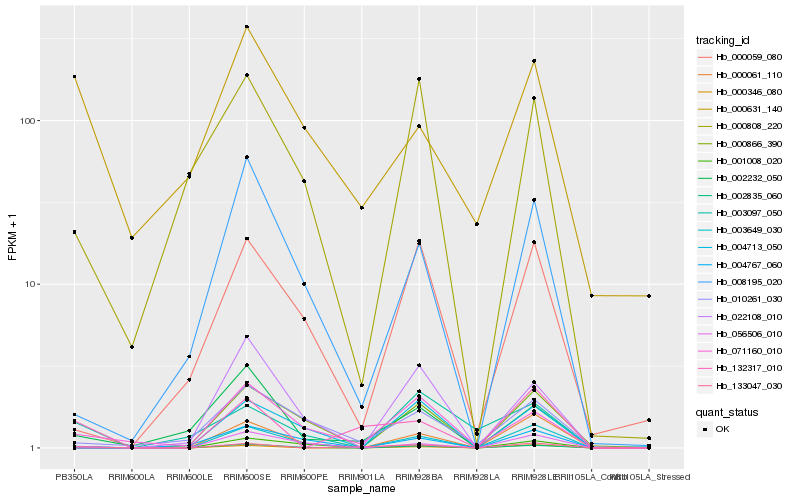
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002835_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640295 [Jatropha curcas] |
| 2 |
Hb_000346_080 |
0.2069117165 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 3 |
Hb_071160_010 |
0.2205789506 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 4 |
Hb_004767_060 |
0.2615265866 |
- |
- |
PREDICTED: uncharacterized protein LOC105761928 [Gossypium raimondii] |
| 5 |
Hb_002232_050 |
0.2711061424 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 6 |
Hb_022108_010 |
0.2716793145 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Glycine soja] |
| 7 |
Hb_000808_220 |
0.2724128839 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 8 |
Hb_133047_030 |
0.2730514336 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_003649_030 |
0.2737821226 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 10 |
Hb_008195_020 |
0.2760633617 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 11 |
Hb_000631_140 |
0.2848449637 |
- |
- |
PREDICTED: chaperone protein ClpB1 [Jatropha curcas] |
| 12 |
Hb_010261_030 |
0.2856317165 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_000866_390 |
0.2892011833 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 14 |
Hb_004713_050 |
0.289916048 |
- |
- |
RecName: Full=Beta-amyrin synthase [Betula platyphylla] |
| 15 |
Hb_000061_110 |
0.2901749592 |
- |
- |
- |
| 16 |
Hb_003097_050 |
0.2920139538 |
- |
- |
hypothetical protein POPTR_0007s07150g [Populus trichocarpa] |
| 17 |
Hb_000059_080 |
0.2953009365 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 18 |
Hb_056506_010 |
0.2990030204 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 19 |
Hb_132317_010 |
0.301659163 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 20 |
Hb_001008_020 |
0.3017705593 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |