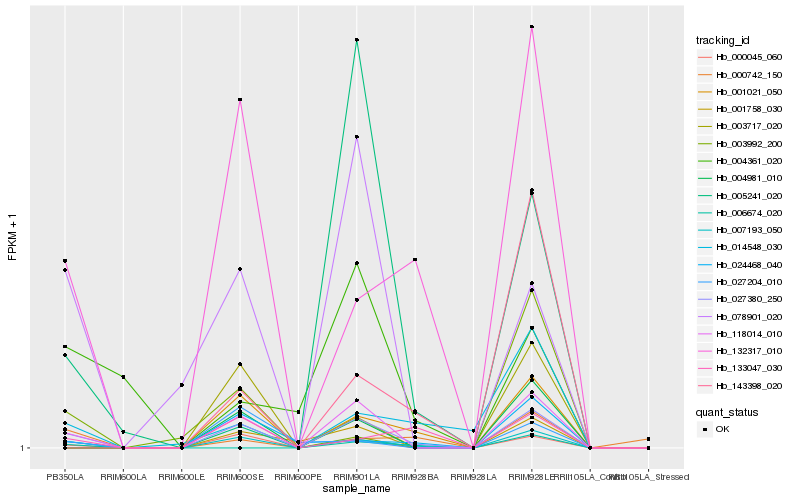
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027380_250 |
0.0 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 2 |
Hb_007193_050 |
0.0824473089 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 3 |
Hb_118014_010 |
0.142125135 |
- |
- |
PREDICTED: uncharacterized protein LOC104214283 [Nicotiana sylvestris] |
| 4 |
Hb_000045_060 |
0.1949532373 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_132317_010 |
0.234271322 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 6 |
Hb_078901_020 |
0.2845221547 |
- |
- |
PREDICTED: uncharacterized protein LOC105648117 [Jatropha curcas] |
| 7 |
Hb_001021_050 |
0.2987812489 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 8 |
Hb_133047_030 |
0.3043805549 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_024468_040 |
0.3089359523 |
- |
- |
- |
| 10 |
Hb_014548_030 |
0.3133671657 |
- |
- |
PREDICTED: uncharacterized protein LOC104897580 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_004981_010 |
0.3134617211 |
- |
- |
- |
| 12 |
Hb_005241_020 |
0.3448635226 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 13 |
Hb_000742_150 |
0.3471186551 |
- |
- |
- |
| 14 |
Hb_003717_020 |
0.3514523964 |
- |
- |
- |
| 15 |
Hb_001758_030 |
0.3542240293 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 16 |
Hb_143398_020 |
0.3544094277 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 17 |
Hb_003992_200 |
0.3586309291 |
- |
- |
- |
| 18 |
Hb_004361_020 |
0.3630268656 |
- |
- |
PREDICTED: uncharacterized protein LOC102625775 [Citrus sinensis] |
| 19 |
Hb_027204_010 |
0.364303299 |
- |
- |
- |
| 20 |
Hb_006674_020 |
0.3643521873 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |