| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
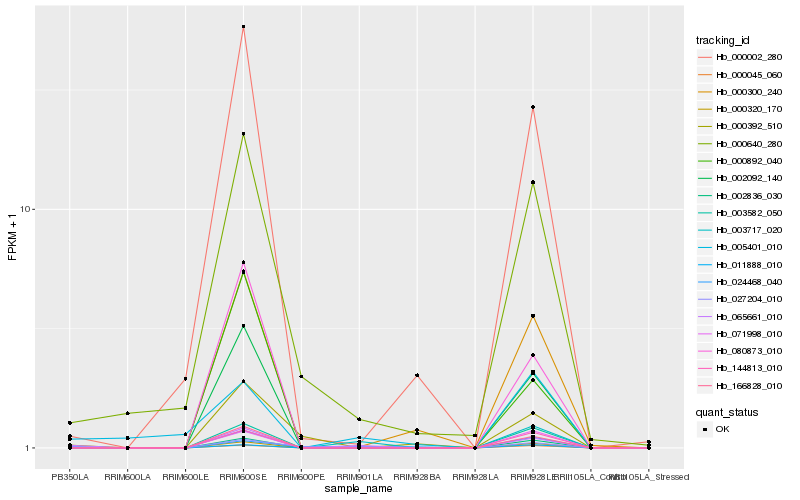
Hb_065661_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 2 |
Hb_027204_010 |
0.1889167447 |
- |
- |
- |
| 3 |
Hb_000045_060 |
0.231309014 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_002092_140 |
0.2697083479 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 5 |
Hb_166828_010 |
0.2700875631 |
- |
- |
- |
| 6 |
Hb_000640_280 |
0.2723873477 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 7 |
Hb_000002_280 |
0.2793509888 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 8 |
Hb_000320_170 |
0.2833079968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_080873_010 |
0.2835307047 |
- |
- |
- |
| 10 |
Hb_003717_020 |
0.284299363 |
- |
- |
- |
| 11 |
Hb_002836_030 |
0.2882246547 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 12 |
Hb_000300_240 |
0.2889728465 |
- |
- |
Sodium/potassium/calcium exchanger 6 precursor, putative [Ricinus communis] |
| 13 |
Hb_003582_050 |
0.2895227355 |
- |
- |
hypothetical protein POPTR_0008s13950g [Populus trichocarpa] |
| 14 |
Hb_144813_010 |
0.2943474722 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 15 |
Hb_000892_040 |
0.3045110621 |
- |
- |
- |
| 16 |
Hb_005401_010 |
0.30702159 |
- |
- |
- |
| 17 |
Hb_011888_010 |
0.3081706519 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 18 |
Hb_024468_040 |
0.3085227105 |
- |
- |
- |
| 19 |
Hb_071998_010 |
0.3100192774 |
- |
- |
PREDICTED: uncharacterized protein LOC105800957 [Gossypium raimondii] |
| 20 |
Hb_000392_510 |
0.315217154 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |