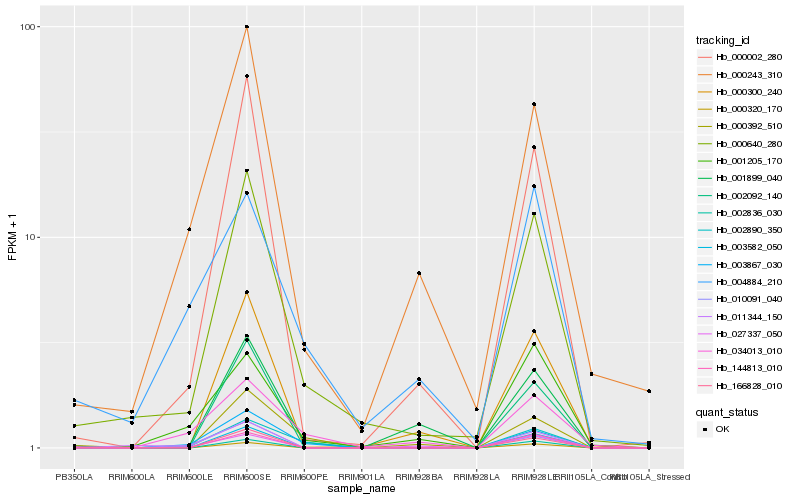
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000640_280 |
0.0 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 2 |
Hb_000002_280 |
0.1760506756 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 3 |
Hb_000243_310 |
0.1919596349 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 4 |
Hb_002890_350 |
0.1921472808 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000300_240 |
0.1982219987 |
- |
- |
Sodium/potassium/calcium exchanger 6 precursor, putative [Ricinus communis] |
| 6 |
Hb_011344_150 |
0.2002985793 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_000392_510 |
0.2047976307 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 8 |
Hb_001899_040 |
0.206081391 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |
| 9 |
Hb_166828_010 |
0.2085480115 |
- |
- |
- |
| 10 |
Hb_000320_170 |
0.2093066955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001205_170 |
0.2096363582 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 12 |
Hb_034013_010 |
0.21050963 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 13 |
Hb_002092_140 |
0.2118163858 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 14 |
Hb_003867_030 |
0.2125930678 |
- |
- |
- |
| 15 |
Hb_002836_030 |
0.2126700646 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 16 |
Hb_003582_050 |
0.2136434354 |
- |
- |
hypothetical protein POPTR_0008s13950g [Populus trichocarpa] |
| 17 |
Hb_027337_050 |
0.2174142544 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 18 |
Hb_144813_010 |
0.2174923358 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 19 |
Hb_004884_210 |
0.2263389536 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 20 |
Hb_010091_040 |
0.2281895373 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |