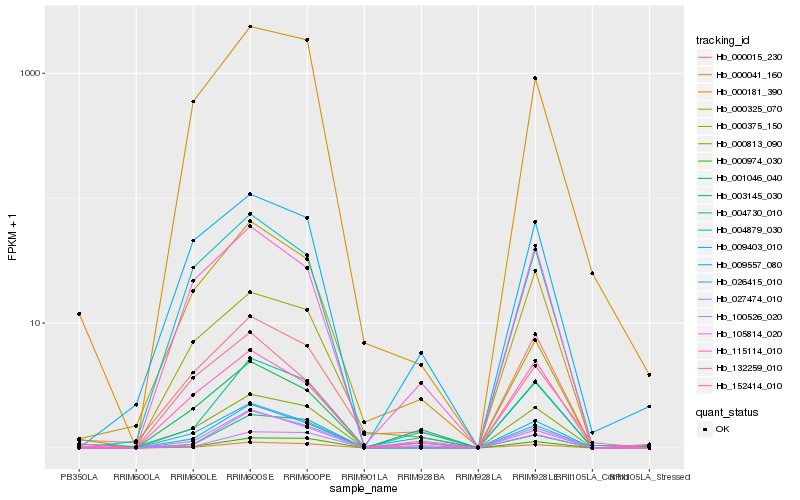
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009403_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 2 |
Hb_026415_010 |
0.0966374199 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004879_030 |
0.1233234246 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 4 |
Hb_000813_090 |
0.1305736546 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 5 |
Hb_000974_030 |
0.1326365 |
- |
- |
PREDICTED: uncharacterized protein LOC105128463 isoform X1 [Populus euphratica] |
| 6 |
Hb_000325_070 |
0.1367732233 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_105814_020 |
0.1385786732 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 8 |
Hb_132259_010 |
0.1432139829 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 9 |
Hb_004730_010 |
0.1433822623 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 10 |
Hb_115114_010 |
0.145969243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000181_390 |
0.1470356429 |
- |
- |
unknown [Lotus japonicus] |
| 12 |
Hb_000015_230 |
0.1531822155 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 13 |
Hb_100526_020 |
0.1584124146 |
- |
- |
hypothetical protein POPTR_0003s09340g [Populus trichocarpa] |
| 14 |
Hb_152414_010 |
0.1591956859 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein PRUPE_ppa024232mg [Prunus persica] |
| 15 |
Hb_001046_040 |
0.1651273638 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_003145_030 |
0.16622662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000041_160 |
0.1703063307 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_009557_080 |
0.1726220665 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 19 |
Hb_000375_150 |
0.1789624531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_027474_010 |
0.1801859603 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |