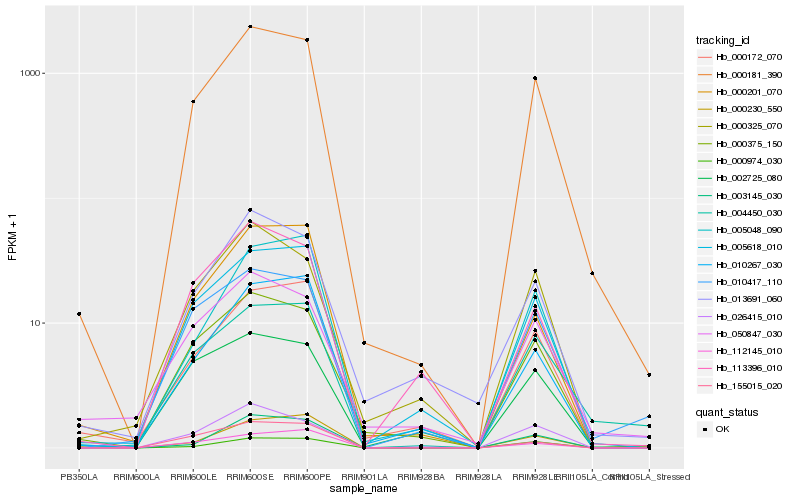
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_390 |
0.0 |
- |
- |
unknown [Lotus japonicus] |
| 2 |
Hb_000375_150 |
0.099134866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005618_010 |
0.1006250225 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 4 |
Hb_026415_010 |
0.1194221256 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004450_030 |
0.1210550089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000172_070 |
0.1220668341 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_010417_110 |
0.1223372927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000325_070 |
0.1296977694 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_000974_030 |
0.1313694497 |
- |
- |
PREDICTED: uncharacterized protein LOC105128463 isoform X1 [Populus euphratica] |
| 10 |
Hb_013691_060 |
0.1338751092 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 11 |
Hb_005048_090 |
0.1349991683 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 12 |
Hb_000201_070 |
0.1359164658 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 13 |
Hb_003145_030 |
0.1374720162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010267_030 |
0.139831751 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_002725_080 |
0.14235201 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 16 |
Hb_050847_030 |
0.143284458 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 17 |
Hb_155015_020 |
0.1436264076 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 18 |
Hb_000230_550 |
0.1454604512 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 19 |
Hb_112145_010 |
0.1461143615 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_113396_010 |
0.147006469 |
- |
- |
cytochrome P450, putative [Ricinus communis] |