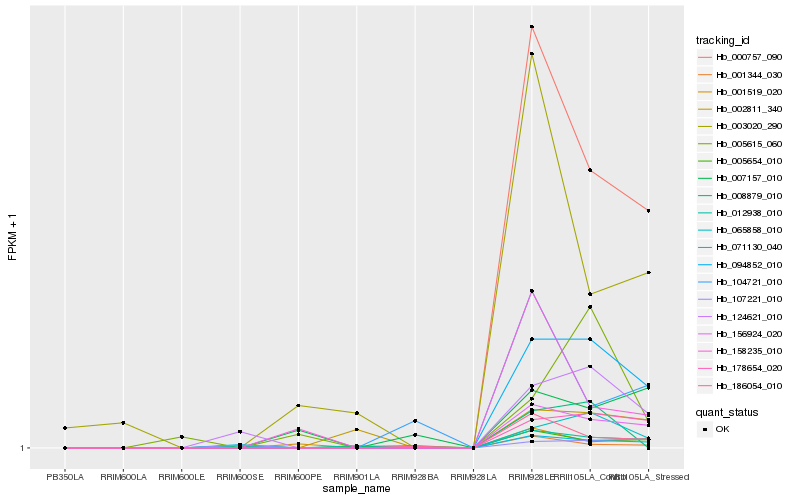
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094852_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 2 |
Hb_156924_020 |
0.0867832249 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 3 |
Hb_178654_020 |
0.1015009723 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 4 |
Hb_000757_090 |
0.1188676453 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 3 [Theobroma cacao] |
| 5 |
Hb_065858_010 |
0.1339806436 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 6 |
Hb_124621_010 |
0.2104536463 |
- |
- |
hypothetical protein L484_019698 [Morus notabilis] |
| 7 |
Hb_001344_030 |
0.2273769901 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_012938_010 |
0.2472264513 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_186054_010 |
0.2555312362 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_071130_040 |
0.2594810183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002811_340 |
0.2901972503 |
- |
- |
PREDICTED: ammonium transporter 3 member 2-like [Populus euphratica] |
| 12 |
Hb_001519_020 |
0.3111822308 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 13 |
Hb_003020_290 |
0.3210209721 |
- |
- |
hypothetical protein CICLE_v10029104mg [Citrus clementina] |
| 14 |
Hb_005615_060 |
0.3236005705 |
- |
- |
PREDICTED: uncharacterized protein LOC105636752 [Jatropha curcas] |
| 15 |
Hb_107221_010 |
0.3242400681 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_104721_010 |
0.3253968348 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 17 |
Hb_158235_010 |
0.3315532782 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 18 |
Hb_005654_010 |
0.3316482365 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_007157_010 |
0.3374687213 |
- |
- |
- |
| 20 |
Hb_008879_010 |
0.3386294777 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |