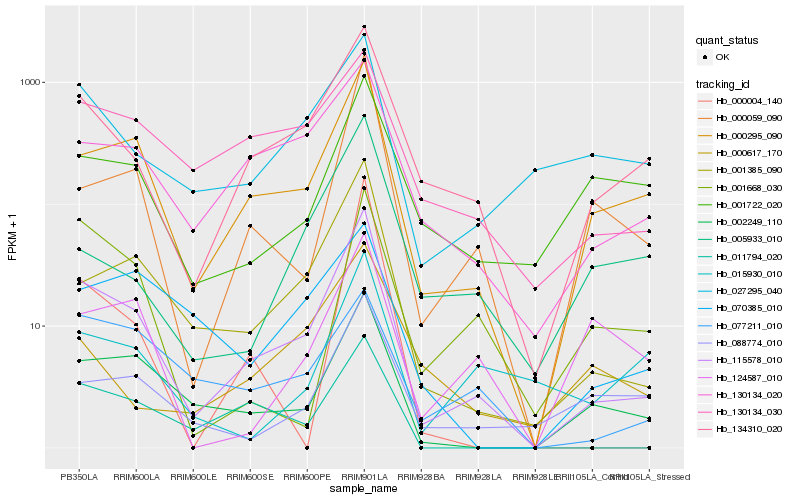
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115578_010 |
0.0 |
- |
- |
PREDICTED: prefoldin subunit 1-like [Populus euphratica] |
| 2 |
Hb_000295_090 |
0.1565610238 |
- |
- |
Clathrin light chain 2 [Glycine soja] |
| 3 |
Hb_001385_090 |
0.1648556145 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 4 |
Hb_134310_020 |
0.1727217432 |
- |
- |
PREDICTED: putative cytochrome c oxidase subunit 5C-4-like [Citrus sinensis] |
| 5 |
Hb_002249_110 |
0.1874886245 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |
| 6 |
Hb_130134_020 |
0.2028316603 |
- |
- |
- |
| 7 |
Hb_000059_090 |
0.2086051768 |
- |
- |
- |
| 8 |
Hb_011794_020 |
0.211507153 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_130134_030 |
0.230103418 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_14472 [Jatropha curcas] |
| 10 |
Hb_088774_010 |
0.2384320227 |
- |
- |
- |
| 11 |
Hb_027295_040 |
0.2401850445 |
- |
- |
hypothetical protein MIMGU_mgv1a017500mg [Erythranthe guttata] |
| 12 |
Hb_000004_140 |
0.2450254235 |
- |
- |
- |
| 13 |
Hb_005933_010 |
0.2491133498 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 14 |
Hb_001722_020 |
0.2521640922 |
- |
- |
- |
| 15 |
Hb_077211_010 |
0.2524529734 |
- |
- |
PREDICTED: lariat debranching enzyme-like [Jatropha curcas] |
| 16 |
Hb_015930_010 |
0.253976228 |
- |
- |
uncharacterized protein LOC100499930 [Glycine max] |
| 17 |
Hb_001668_030 |
0.2559733923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_070385_010 |
0.2598727281 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 19 |
Hb_000617_170 |
0.2599053554 |
- |
- |
- |
| 20 |
Hb_124587_010 |
0.2599358629 |
- |
- |
PREDICTED: importin-5-like [Musa acuminata subsp. malaccensis] |