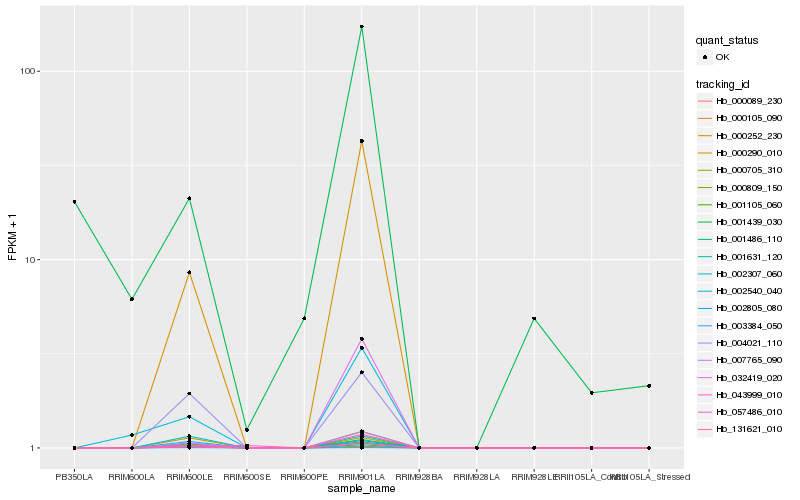
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000809_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_001105_060 |
0.0024418991 |
- |
- |
hypothetical protein CISIN_1g0045352mg, partial [Citrus sinensis] |
| 3 |
Hb_000252_230 |
0.027171784 |
- |
- |
hypothetical protein B456_008G160800 [Gossypium raimondii] |
| 4 |
Hb_043999_010 |
0.0462127203 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 5 |
Hb_007765_090 |
0.0480838083 |
- |
- |
unknown [Glycine max] |
| 6 |
Hb_131621_010 |
0.0726970919 |
- |
- |
hypothetical protein MIMGU_mgv1a004274mg [Erythranthe guttata] |
| 7 |
Hb_001486_110 |
0.1210945841 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |
| 8 |
Hb_002540_040 |
0.1703106774 |
- |
- |
BnaC05g39550D [Brassica napus] |
| 9 |
Hb_004021_110 |
0.1964522238 |
- |
- |
- |
| 10 |
Hb_032419_020 |
0.2603929939 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 11 |
Hb_000705_310 |
0.260628228 |
- |
- |
hypothetical protein POPTR_0001s03390g [Populus trichocarpa] |
| 12 |
Hb_001631_120 |
0.2616126887 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_000105_090 |
0.2628293838 |
- |
- |
- |
| 14 |
Hb_002805_080 |
0.2645392554 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 15 |
Hb_003384_050 |
0.2702715353 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_000089_230 |
0.271325285 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 17 |
Hb_057486_010 |
0.2715253174 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |
| 18 |
Hb_000290_010 |
0.2858655639 |
- |
- |
- |
| 19 |
Hb_002307_060 |
0.2887418885 |
- |
- |
- |
| 20 |
Hb_001439_030 |
0.2966095186 |
- |
- |
- |