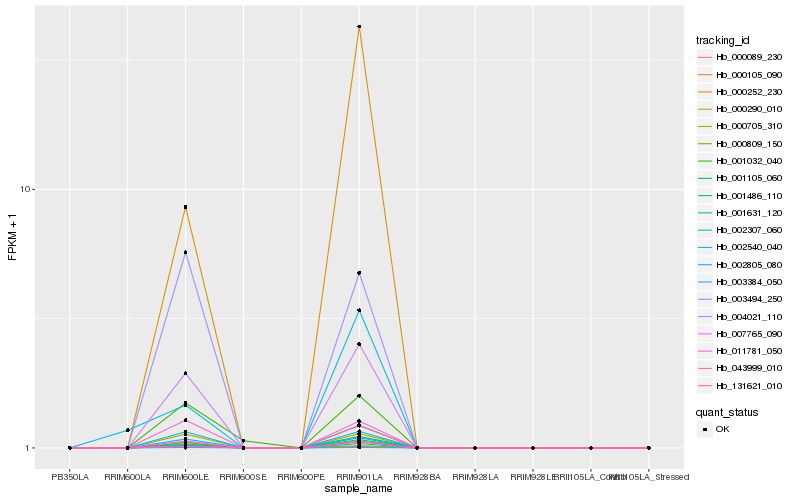
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_110 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |
| 2 |
Hb_131621_010 |
0.0486566909 |
- |
- |
hypothetical protein MIMGU_mgv1a004274mg [Erythranthe guttata] |
| 3 |
Hb_007765_090 |
0.0732789423 |
- |
- |
unknown [Glycine max] |
| 4 |
Hb_043999_010 |
0.0751469377 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 5 |
Hb_004021_110 |
0.0762795736 |
- |
- |
- |
| 6 |
Hb_001105_060 |
0.1186764043 |
- |
- |
hypothetical protein CISIN_1g0045352mg, partial [Citrus sinensis] |
| 7 |
Hb_000809_150 |
0.1210945841 |
- |
- |
- |
| 8 |
Hb_000705_310 |
0.1416313408 |
- |
- |
hypothetical protein POPTR_0001s03390g [Populus trichocarpa] |
| 9 |
Hb_001631_120 |
0.142636221 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000105_090 |
0.1438782459 |
- |
- |
- |
| 11 |
Hb_002805_080 |
0.1456238886 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 12 |
Hb_000252_230 |
0.1479209431 |
- |
- |
hypothetical protein B456_008G160800 [Gossypium raimondii] |
| 13 |
Hb_003384_050 |
0.1514775619 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_000089_230 |
0.1525538722 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 15 |
Hb_000290_010 |
0.1674131025 |
- |
- |
- |
| 16 |
Hb_002307_060 |
0.1703541883 |
- |
- |
- |
| 17 |
Hb_011781_050 |
0.1835220056 |
- |
- |
- |
| 18 |
Hb_002540_040 |
0.2174542437 |
- |
- |
BnaC05g39550D [Brassica napus] |
| 19 |
Hb_001032_040 |
0.2176959622 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003494_250 |
0.2229241841 |
- |
- |
- |