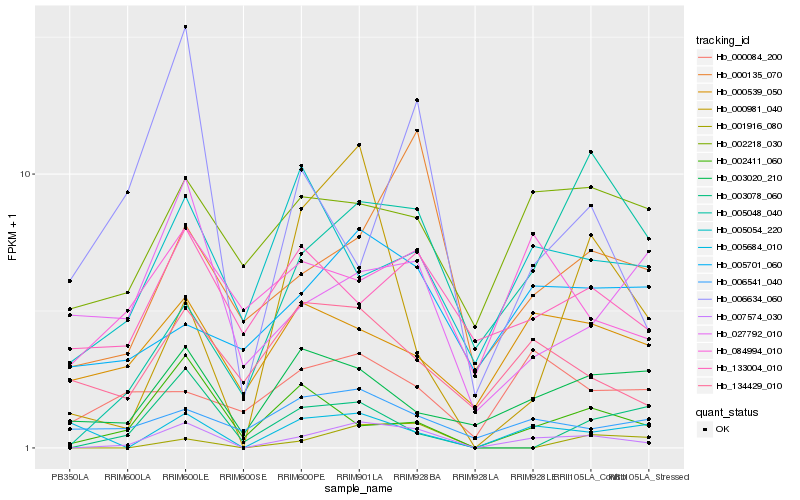
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007574_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s24510g [Populus trichocarpa] |
| 2 |
Hb_000135_070 |
0.3083020532 |
- |
- |
PREDICTED: inositol oxygenase 1-like [Jatropha curcas] |
| 3 |
Hb_001916_080 |
0.3204169285 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 4 |
Hb_002411_060 |
0.3215600721 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 5 |
Hb_003020_210 |
0.326671661 |
- |
- |
Structural maintenance of chromosomes 2-2 -like protein [Gossypium arboreum] |
| 6 |
Hb_134429_010 |
0.3278490366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005048_040 |
0.3283204363 |
- |
- |
PREDICTED: endonuclease 1 [Jatropha curcas] |
| 8 |
Hb_006541_040 |
0.3284738441 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 2 [Jatropha curcas] |
| 9 |
Hb_005701_060 |
0.3325523174 |
- |
- |
hypothetical protein JCGZ_04513 [Jatropha curcas] |
| 10 |
Hb_002218_030 |
0.3341142509 |
- |
- |
hypothetical protein B456_007G345200 [Gossypium raimondii] |
| 11 |
Hb_000539_050 |
0.3353555832 |
- |
- |
bile acid:sodium symporter, putative [Ricinus communis] |
| 12 |
Hb_084994_010 |
0.3362876621 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005684_010 |
0.3376513325 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 14 |
Hb_027792_010 |
0.3418223955 |
- |
- |
- |
| 15 |
Hb_133004_010 |
0.3423238645 |
- |
- |
PREDICTED: cyclin-D3-3 [Jatropha curcas] |
| 16 |
Hb_005054_220 |
0.3431170442 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 17 |
Hb_003078_060 |
0.3435014387 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 18 |
Hb_000084_200 |
0.3447723524 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X2 [Jatropha curcas] |
| 19 |
Hb_006634_060 |
0.3453760761 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 20 |
Hb_000981_040 |
0.348410392 |
- |
- |
- |