| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
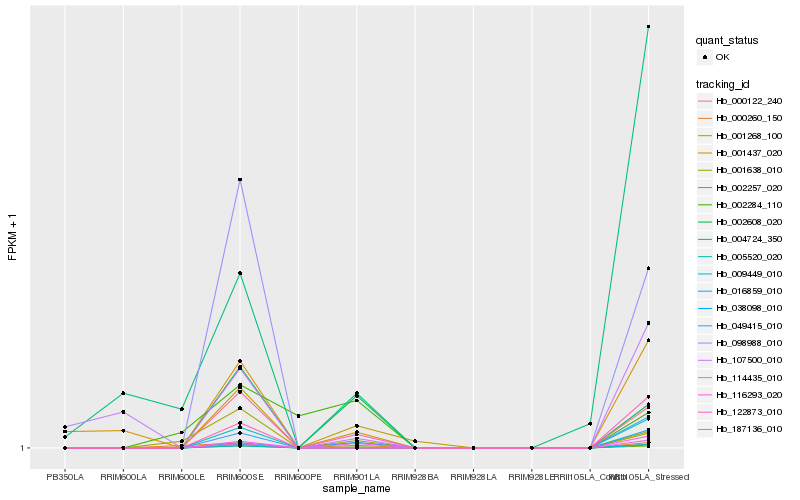
Hb_002257_020 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 2 |
Hb_002608_020 |
0.1466089862 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 305 [Jatropha curcas] |
| 3 |
Hb_114435_010 |
0.1509653127 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 4 |
Hb_122873_010 |
0.1717980188 |
- |
- |
- |
| 5 |
Hb_000122_240 |
0.2076965743 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 6 |
Hb_038098_010 |
0.2465150695 |
- |
- |
PREDICTED: uncharacterized protein LOC105793140 [Gossypium raimondii] |
| 7 |
Hb_001437_020 |
0.2805708528 |
- |
- |
PREDICTED: uncharacterized protein LOC105628060 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000260_150 |
0.2875523142 |
- |
- |
hypothetical protein POPTR_0344s002001g, partial [Populus trichocarpa] |
| 9 |
Hb_001638_010 |
0.2896377968 |
- |
- |
- |
| 10 |
Hb_001268_100 |
0.3041921433 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 11 |
Hb_016859_010 |
0.3183474275 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_187136_010 |
0.3221171783 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 13 |
Hb_009449_010 |
0.3316619161 |
- |
- |
Serine/threonine-protein kinase HT1 [Gossypium arboreum] |
| 14 |
Hb_098988_010 |
0.3325024032 |
- |
- |
hypothetical protein JCGZ_18539 [Jatropha curcas] |
| 15 |
Hb_116293_020 |
0.3462194006 |
- |
- |
Uncharacterized protein TCM_043787 [Theobroma cacao] |
| 16 |
Hb_005520_020 |
0.3466354962 |
- |
- |
hypothetical protein EUGRSUZ_G03336 [Eucalyptus grandis] |
| 17 |
Hb_049415_010 |
0.3605149212 |
- |
- |
- |
| 18 |
Hb_002284_110 |
0.390495716 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP18, chloroplastic isoform X2 [Solanum lycopersicum] |
| 19 |
Hb_107500_010 |
0.3933294978 |
- |
- |
- |
| 20 |
Hb_004724_350 |
0.398582881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |